Sexual Dimorphism and Aging in the Human Hyppocampus: Identification, Validation, and Impact of Differentially Expressed Genes by Factorial Microarray and Network Analysis
- PMID: 27761111
- PMCID: PMC5050216
- DOI: 10.3389/fnagi.2016.00229
Sexual Dimorphism and Aging in the Human Hyppocampus: Identification, Validation, and Impact of Differentially Expressed Genes by Factorial Microarray and Network Analysis
Abstract
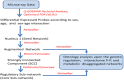
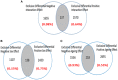
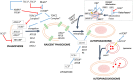
Motivation: In the brain of elderly-healthy individuals, the effects of sexual dimorphism and those due to normal aging appear overlapped. Discrimination of these two dimensions would powerfully contribute to a better understanding of the etiology of some neurodegenerative diseases, such as "sporadic" Alzheimer. Methods: Following a system biology approach, top-down and bottom-up strategies were combined. First, public transcriptome data corresponding to the transition from adulthood to the aging stage in normal, human hippocampus were analyzed through an optimized microarray post-processing (Q-GDEMAR method) together with a proper experimental design (full factorial analysis). Second, the identified genes were placed in context by building compatible networks. The subsequent ontology analyses carried out on these networks clarify the main functionalities involved. Results: Noticeably we could identify large sets of genes according to three groups: those that exclusively depend on the sex, those that exclusively depend on the age, and those that depend on the particular combinations of sex and age (interaction). The genes identified were validated against three independent sources (a proteomic study of aging, a senescence database, and a mitochondrial genetic database). We arrived to several new inferences about the biological functions compromised during aging in two ways: by taking into account the sex-independent effects of aging, and considering the interaction between age and sex where pertinent. In particular, we discuss the impact of our findings on the functions of mitochondria, autophagy, mitophagia, and microRNAs. Conclusions: The evidence obtained herein supports the occurrence of significant neurobiological differences in the hippocampus, not only between adult and elderly individuals, but between old-healthy women and old-healthy men. Hence, to obtain realistic results in further analysis of the transition from the normal aging to incipient Alzheimer, the features derived from the sexual dimorphism in hippocampus should be explicitly considered.
Keywords: aging; autophagia; hippocampus; microRNAs; microarray; mitochondria; senescence; sexual differences.
Figures


Similar articles
-
Harnessing the power of gene microarrays for the study of brain aging and Alzheimer's disease: statistical reliability and functional correlation.Ageing Res Rev. 2005 Nov;4(4):481-512. doi: 10.1016/j.arr.2005.06.006. Epub 2005 Oct 27. Ageing Res Rev. 2005. PMID: 16257272 Review.
-
Co-expression network analysis identified hub genes critical to triglyceride and free fatty acid metabolism as key regulators of age-related vascular dysfunction in mice.Aging (Albany NY). 2019 Sep 12;11(18):7620-7638. doi: 10.18632/aging.102275. Epub 2019 Sep 12. Aging (Albany NY). 2019. PMID: 31514170 Free PMC article.
-
Q-GDEMAR: a general method for the identification of differentially expressed genes in microarrays with unbalanced groups.Mol Biosyst. 2016 Jan;12(1):120-32. doi: 10.1039/c5mb00541h. Mol Biosyst. 2016. PMID: 26563436
-
Influence of Glucose Availability and CRP Acetylation on the Genome-Wide Transcriptional Response of Escherichia coli: Assessment by an Optimized Factorial Microarray Analysis.Front Microbiol. 2018 May 23;9:941. doi: 10.3389/fmicb.2018.00941. eCollection 2018. Front Microbiol. 2018. PMID: 29875739 Free PMC article.
-
MitomiRs in human inflamm-aging: a hypothesis involving miR-181a, miR-34a and miR-146a.Exp Gerontol. 2014 Aug;56:154-63. doi: 10.1016/j.exger.2014.03.002. Epub 2014 Mar 7. Exp Gerontol. 2014. PMID: 24607549 Review.
Cited by
-
Age-related neurodegenerative diseases.J Cell Physiol. 2020 Apr;235(4):3131-3141. doi: 10.1002/jcp.29248. Epub 2019 Sep 25. J Cell Physiol. 2020. PMID: 31556109 Free PMC article. Review.
-
Influence of Normal Aging on Brain Autophagy: A Complex Scenario.Front Aging Neurosci. 2019 Mar 11;11:49. doi: 10.3389/fnagi.2019.00049. eCollection 2019. Front Aging Neurosci. 2019. PMID: 30914945 Free PMC article. Review.
-
Possible sexually dimorphic role of miRNA and other sncRNA in ASD brain.Mol Autism. 2017 Feb 7;8:4. doi: 10.1186/s13229-017-0117-0. eCollection 2017. Mol Autism. 2017. PMID: 28184278 Free PMC article.
-
Mapping the transcriptomic changes of endothelial compartment in human hippocampus across aging and mild cognitive impairment.Biol Open. 2021 May 15;10(5):bio057950. doi: 10.1242/bio.057950. Epub 2021 May 17. Biol Open. 2021. PMID: 34184731 Free PMC article.
-
Role of Sex Hormones on Brain Mitochondrial Function, with Special Reference to Aging and Neurodegenerative Diseases.Front Aging Neurosci. 2017 Dec 7;9:406. doi: 10.3389/fnagi.2017.00406. eCollection 2017. Front Aging Neurosci. 2017. PMID: 29270123 Free PMC article. Review.
References
-
- Baj G., Del Turco D., Schlaudraff J., Torelli L., Deller T., Tongiorgi E. (2013). Regulation of the spatial code for BDNF mRNA isoforms in the rat hippocampus following pilocarpine-treatment: a systematic analysis using laser microdissection and quantitative real-time PCR. Hippocampus 23, 413–423. 10.1002/hipo.22100 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

