Identification and temporal expression of putative circadian clock transcripts in the amphipod crustacean Talitrus saltator
- PMID: 27761341
- PMCID: PMC5068443
- DOI: 10.7717/peerj.2555
Identification and temporal expression of putative circadian clock transcripts in the amphipod crustacean Talitrus saltator
Abstract
Background: Talitrus saltator is an amphipod crustacean that inhabits the supralittoral zone on sandy beaches in the Northeast Atlantic and Mediterranean. T. saltator exhibits endogenous locomotor activity rhythms and time-compensated sun and moon orientation, both of which necessitate at least one chronometric mechanism. Whilst their behaviour is well studied, currently there are no descriptions of the underlying molecular components of a biological clock in this animal, and very few in other crustacean species.
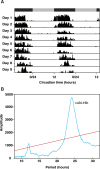
Methods: We harvested brain tissue from animals expressing robust circadian activity rhythms and used homology cloning and Illumina RNAseq approaches to sequence and identify the core circadian clock and clock-related genes in these samples. We assessed the temporal expression of these genes in time-course samples from rhythmic animals using RNAseq.
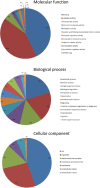
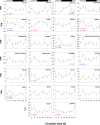
Results: We identified a comprehensive suite of circadian clock gene homologues in T. saltator including the 'core' clock genes period (Talper), cryptochrome 2 (Talcry2), timeless (Taltim), clock (Talclk), and bmal1 (Talbmal1). In addition we describe the sequence and putative structures of 23 clock-associated genes including two unusual, extended isoforms of pigment dispersing hormone (Talpdh). We examined time-course RNAseq expression data, derived from tissues harvested from behaviourally rhythmic animals, to reveal rhythmic expression of these genes with approximately circadian period in Talper and Talbmal1. Of the clock-related genes, casein kinase IIβ (TalckIIβ), ebony (Talebony), jetlag (Taljetlag), pigment dispensing hormone (Talpdh), protein phosphatase 1 (Talpp1), shaggy (Talshaggy), sirt1 (Talsirt1), sirt7 (Talsirt7) and supernumerary limbs (Talslimb) show temporal changes in expression.
Discussion: We report the sequences of principle genes that comprise the circadian clock of T. saltator and highlight the conserved structural and functional domains of their deduced cognate proteins. Our sequencing data contribute to the growing inventory of described comparative clocks. Expression profiling of the identified clock genes illuminates tantalising targets for experimental manipulation to elucidate the molecular and cellular control of clock-driven phenotypes in this crustacean.
Keywords: Circadian; Crustacean; Rhythms; Talitrus saltator; Transcriptome.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, De Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, Mcintosh TC, Mcleod MP, Mcpherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

