Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome
- PMID: 27761921
- PMCID: PMC5177492
- DOI: 10.1002/ana.24800
Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome
Abstract
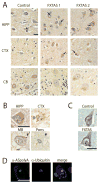
Objective: Repeat-associated non-AUG (RAN) translation drives production of toxic proteins from pathogenic repeat sequences in multiple untreatable neurodegenerative disorders. Fragile X-associated tremor/ataxia syndrome (FXTAS) is one such condition, resulting from a CGG trinucleotide repeat expansion in the 5' leader sequence of the FMR1 gene. RAN proteins from the CGG repeat accumulate in ubiquitinated inclusions in FXTAS patient brains and elicit toxicity. In addition to the CGG repeat, an antisense mRNA containing a CCG repeat is also transcribed from the FMR1 locus. We evaluated whether this antisense CCG repeat supports RAN translation and contributes to pathology in FXTAS patients.
Methods: We generated a series of CCG RAN translation-specific reporters and utilized them to measure RAN translation from CCG repeats in multiple reading frames in transfected cells. We also developed antibodies against predicted CCG RAN proteins and used immunohistochemistry and immunofluorescence on FXTAS patient tissues to measure their accumulation and distribution.
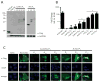
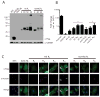
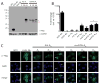
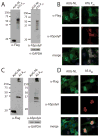
Results: RAN translation from CCG repeats is supported in all 3 potential reading frames, generating polyproline, polyarginine, and polyalanine proteins, respectively. Their production occurs whether or not the natural AUG start upstream of the repeat in the proline reading frame is present. All 3 frames show greater translation at larger repeat sizes. Antibodies targeted to the antisense FMR polyproline and polyalanine proteins selectively stain nuclear and cytoplasmic aggregates in FXTAS patients and colocalize with ubiquitinated neuronal inclusions.
Interpretation: RAN translation from antisense CCG repeats generates novel proteins that accumulate in ubiquitinated inclusions in FXTAS patients. Ann Neurol 2016;80:871-881.
© 2016 American Neurological Association.
Conflict of interest statement
POTENTIAL CONFLICTS OF INTEREST All authors declare no conflicts of interest.
Figures


Similar articles
-
Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome.Brain Res. 2018 Aug 15;1693(Pt A):43-54. doi: 10.1016/j.brainres.2018.02.006. Epub 2018 Feb 14. Brain Res. 2018. PMID: 29453961 Free PMC article. Review.
-
Neuropathology of RAN translation proteins in fragile X-associated tremor/ataxia syndrome.Acta Neuropathol Commun. 2019 Oct 30;7(1):152. doi: 10.1186/s40478-019-0782-7. Acta Neuropathol Commun. 2019. PMID: 31665086 Free PMC article.
-
CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome.Neuron. 2013 May 8;78(3):440-55. doi: 10.1016/j.neuron.2013.03.026. Epub 2013 Apr 18. Neuron. 2013. PMID: 23602499 Free PMC article.
-
Astroglial-targeted expression of the fragile X CGG repeat premutation in mice yields RAN translation, motor deficits and possible evidence for cell-to-cell propagation of FXTAS pathology.Acta Neuropathol Commun. 2019 Feb 26;7(1):27. doi: 10.1186/s40478-019-0677-7. Acta Neuropathol Commun. 2019. PMID: 30808398 Free PMC article.
-
Potential pathogenic mechanisms underlying Fragile X Tremor Ataxia Syndrome: RAN translation and/or RNA gain-of-function?Eur J Med Genet. 2018 Nov;61(11):674-679. doi: 10.1016/j.ejmg.2017.11.001. Epub 2017 Dec 6. Eur J Med Genet. 2018. PMID: 29223504 Review.
Cited by
-
DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation.EMBO Rep. 2019 Sep;20(9):e47498. doi: 10.15252/embr.201847498. Epub 2019 Jul 25. EMBO Rep. 2019. PMID: 31347257 Free PMC article.
-
Fragile X-Associated Tremor/Ataxia Syndrome (FXTAS): A Gender Perspective.J Clin Med. 2022 Feb 15;11(4):1002. doi: 10.3390/jcm11041002. J Clin Med. 2022. PMID: 35207276 Free PMC article. Review.
-
Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome.Brain Res. 2018 Aug 15;1693(Pt A):43-54. doi: 10.1016/j.brainres.2018.02.006. Epub 2018 Feb 14. Brain Res. 2018. PMID: 29453961 Free PMC article. Review.
-
New developments in RAN translation: insights from multiple diseases.Curr Opin Genet Dev. 2017 Jun;44:125-134. doi: 10.1016/j.gde.2017.03.006. Epub 2017 Mar 30. Curr Opin Genet Dev. 2017. PMID: 28365506 Free PMC article. Review.
-
Neuropathology of RAN translation proteins in fragile X-associated tremor/ataxia syndrome.Acta Neuropathol Commun. 2019 Oct 30;7(1):152. doi: 10.1186/s40478-019-0782-7. Acta Neuropathol Commun. 2019. PMID: 31665086 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

