A unique peptide deformylase platform to rationally design and challenge novel active compounds
- PMID: 27762275
- PMCID: PMC5071857
- DOI: 10.1038/srep35429
A unique peptide deformylase platform to rationally design and challenge novel active compounds
Erratum in
-
Corrigendum: A unique peptide deformylase platform to rationally design and challenge novel active compounds.Sci Rep. 2017 Jan 11;7:39365. doi: 10.1038/srep39365. Sci Rep. 2017. PMID: 28074917 Free PMC article. No abstract available.
Abstract
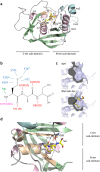
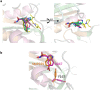
Peptide deformylase (PDF) is considered an excellent target to develop antibiotics. We have performed an extensive characterization of a new PDF from the pathogen Streptococcus agalactiae, showing properties similar to other known PDFs. S. agalactiae PDF could be used as PDF prototype as it allowed to get complete sets of 3-dimensional, biophysical and kinetic data with virtually any inhibitor compound. Structure-activity relationship analysis with this single reference system allowed us to reveal distinct binding modes for different PDF inhibitors and the key role of a hydrogen bond in potentiating the interaction between ligand and target. We propose this protein as an irreplaceable tool, allowing easy and relevant fine comparisons between series, to design, challenge and validate novel series of inhibitors. As proof-of-concept, we report here the design and synthesis of effective specific bacterial PDF inhibitors of an oxadiazole series with potent antimicrobial activity against a multidrug resistant clinical isolate.
Figures


Similar articles
-
Peptide deformylases from Vibrio parahaemolyticus phage and bacteria display similar deformylase activity and inhibitor binding clefts.Biochim Biophys Acta Proteins Proteom. 2018 Feb;1866(2):348-355. doi: 10.1016/j.bbapap.2017.10.007. Epub 2017 Oct 31. Biochim Biophys Acta Proteins Proteom. 2018. PMID: 29101077
-
The evolution of peptide deformylase as a target: contribution of biochemistry, genetics and genomics.Biochem Pharmacol. 2006 Mar 30;71(7):1042-7. doi: 10.1016/j.bcp.2005.10.015. Epub 2005 Nov 11. Biochem Pharmacol. 2006. PMID: 16289392 Review.
-
Characterization of a human peptide deformylase: implications for antibacterial drug design.Biochemistry. 2003 Aug 26;42(33):9952-8. doi: 10.1021/bi0346446. Biochemistry. 2003. PMID: 12924944
-
Bacterial Peptide Deformylase Inhibition of Tetrazole-Substituted Biaryl Acid Analogs: Synthesis, Biological Evaluations, and Molecular Docking Study.Arch Pharm (Weinheim). 2016 Dec;349(12):934-943. doi: 10.1002/ardp.201600254. Epub 2016 Nov 16. Arch Pharm (Weinheim). 2016. PMID: 27859538
-
Novel approaches to antimicrobial therapy: peptide deformylase.Curr Opin Drug Discov Devel. 2002 Sep;5(5):785-92. Curr Opin Drug Discov Devel. 2002. PMID: 12630299 Review.
Cited by
-
Total synthesis of (±)-fumimycin and analogues for biological evaluation as peptide deformylase inhibitors.Tetrahedron. 2019 Jun 14;75(24):3216-3230. doi: 10.1016/j.tet.2019.03.037. Epub 2019 Mar 27. Tetrahedron. 2019. PMID: 31555018 Free PMC article.
-
Prediction of Pharmacokinetics of IDP-73152 in Humans Using Physiologically-Based Pharmacokinetics.Pharmaceutics. 2022 May 28;14(6):1157. doi: 10.3390/pharmaceutics14061157. Pharmaceutics. 2022. PMID: 35745730 Free PMC article.
-
Data on molecular docking of naturally occurring flavonoids with biologically important targets.Data Brief. 2020 Feb 4;29:105243. doi: 10.1016/j.dib.2020.105243. eCollection 2020 Apr. Data Brief. 2020. PMID: 32072001 Free PMC article.
-
Metal utilization in genome-reduced bacteria: Do human mycoplasmas rely on iron?Comput Struct Biotechnol J. 2021 Oct 18;19:5752-5761. doi: 10.1016/j.csbj.2021.10.022. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34765092 Free PMC article. Review.
-
Discovery of Potential Plant-Derived Peptide Deformylase (PDF) Inhibitors for Multidrug-Resistant Bacteria Using Computational Studies.J Clin Med. 2018 Dec 17;7(12):563. doi: 10.3390/jcm7120563. J Clin Med. 2018. PMID: 30563019 Free PMC article.
References
-
- Brown D. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov 14, 821–832 (2015). - PubMed
-
- Berendonk T. U. et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13, 310–317 (2015). - PubMed
-
- Tommasi R., Brown D. G., Walkup G. K., Manchester J. I. & Miller A. A. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14, 529–542 (2015). - PubMed
-
- Laxminarayan R. et al. Access to effective antimicrobials: a worldwide challenge. Lancet 387, 168–175 (2016). - PubMed
-
- Drebes J., Kunz M., Pereira C. A., Betzel C. & Wrenger C. MRSA infections: from classical treatment to suicide drugs. Curr Med Chem 21, 1809–1819 (2014). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

