Hepatitis C RNA assay differences in results: Potential implications for shortened therapy and determination of Sustained Virologic Response
- PMID: 27762283
- PMCID: PMC5071881
- DOI: 10.1038/srep35410
Hepatitis C RNA assay differences in results: Potential implications for shortened therapy and determination of Sustained Virologic Response
Abstract
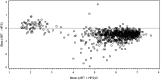
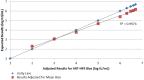
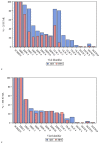
Approval of Ledipasvir/Sofosbuvir for the treatment of chronic hepatitis C (HCV) includes the truncation of therapy from 12 to 8 weeks in treatment naïve, non-cirrhotic patients with baseline HCV RNA levels <6 million IU/mL (6.8 log10 IU/mL). The aim of this study was to evaluate this clinical cutoff with a different widely used commercially available HCV RNA test. Results from samples tested prospectively with Roche High Pure TaqMan HCV 2.0 test (HPS) were compared to those tested retrospectively with the Abbott RealTime HCV RNA test (ART). Using 6 million IU/mL as the cut-off, pre-treatment results were concordant in 70.4% of cases. When results with the same test measured at screening and baseline, clinical decisions could be impacted in 14.4% and 6.2% of cases for HPS and ART respectively. Using only HCV RNA cutoff of 6 million IU/mL, 29.55% of subjects would receive a different and potentially incorrect treatment duration based solely on HCV RNA test method used. A further 6-14% of subjects would have treatment decision change based on the day the sample was taken.
Trial registration: ClinicalTrials.gov NCT01716585.
Conflict of interest statement
Gavin Cloherty, Kevin Cheng, Christine Herman, Vera Holzmayer, George Dawson and John Hackett are employees and shareholders of Abbott Laboratories. Heiner Wedemeyer has received honoraria for consulting or speaking engagements from Abbott, Abbvie, Biolex, Bristol-Myers Squibb, Boehringer Ingelheim, Eiger Pharmaceuticals, Falk Foundation, Gilead Sciences, ITS, JJ/Janssen-Cilag/Janssen TE, Medgenics, Merck/ Schering-Plough, Novartis, Novaria, Roche, Roche Diagnostics, Siemens, Transgene and ViiV. Jean-Michel Pawlotsky has received research grants from Gilead Sciences and has served as an advisor fror Abbott, Abbvie, Achillon, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Idenix, Janssen, Merck, Novartis and Roche. Christoph Sarrazin has received speaker fees, advisory boards and research support from Abbvie, Roche, Siemens and Qiagen. Stephen Chevaliez has received research grants from Gilead Sciences and has served as an advisor for Gilead Sciences and Roche Pharmaceuticals. Jordan Feld has received consulting fees from Abbvie, Abbott, BMS, Gilead, Janssen, Merck, Theravance. Research support from Abbvie, Abbott, BI, BMS, Gilead, Janssen, Merck, Santaris. Benjamin Maasoumy has received speaker and/or consulting fees from Abbott Molecular, Roche, MSD/Merck, BMS, Fujirebio, Janssen-Cilaq. Research support from Roche and Abbott Molecular. Travel grants from Janssen-Cilaq and Gilead. Johannes Vermehren has received consulting and lecture fees from Abbott, AbbVie, Bristol-Myers Squibb, Covidien, Gilead.
Figures



Similar articles
-
An OPTIMIZE study retrospective analysis for management of telaprevir-treated hepatitis C virus (HCV)-infected patients by use of the Abbott RealTime HCV RNA assay.J Clin Microbiol. 2015 Apr;53(4):1264-9. doi: 10.1128/JCM.03030-14. Epub 2015 Feb 4. J Clin Microbiol. 2015. PMID: 25653396 Free PMC article.
-
Comparison of on-treatment HCV RNA during direct antiviral therapy using two different COBAS TaqMan HCV assays.J Clin Virol. 2017 Apr;89:51-56. doi: 10.1016/j.jcv.2017.02.008. Epub 2017 Feb 24. J Clin Virol. 2017. PMID: 28259054
-
HCV RNA quantification with different assays: implications for protease-inhibitor-based response-guided therapy.Antivir Ther. 2014;19(6):559-67. doi: 10.3851/IMP2760. Epub 2014 Feb 28. Antivir Ther. 2014. PMID: 24584086
-
Applicability of Hepatitis C Virus RNA Viral Load Thresholds for 8-Week Treatments in Patients With Chronic Hepatitis C Virus Genotype 1 Infection.Clin Infect Dis. 2016 May 15;62(10):1228-1234. doi: 10.1093/cid/ciw061. Epub 2016 Feb 9. Clin Infect Dis. 2016. PMID: 26908802
-
Fixed-dose combination of sofosbuvir and ledipasvir for the treatment of chronic hepatitis C genotype 1.Expert Opin Pharmacother. 2015 Apr;16(5):739-48. doi: 10.1517/14656566.2015.1013938. Epub 2015 Feb 13. Expert Opin Pharmacother. 2015. PMID: 25676581 Review.
Cited by
-
HCV-Specific T Cell Responses During and After Chronic HCV Infection.Viruses. 2018 Nov 17;10(11):645. doi: 10.3390/v10110645. Viruses. 2018. PMID: 30453612 Free PMC article. Review.
-
Effect of Low Positive End of Treatment Viral Load with Direct-Acting Antiviral Therapy on Sustained Virologic Response.Can J Gastroenterol Hepatol. 2020 Jul 27;2020:8815829. doi: 10.1155/2020/8815829. eCollection 2020. Can J Gastroenterol Hepatol. 2020. PMID: 32802821 Free PMC article.
-
Decrease of T-cells exhaustion markers programmed cell death-1 and T-cell immunoglobulin and mucin domain-containing protein 3 and plasma IL-10 levels after successful treatment of chronic hepatitis C.Sci Rep. 2020 Sep 29;10(1):16060. doi: 10.1038/s41598-020-73137-6. Sci Rep. 2020. PMID: 32994477 Free PMC article.
References
-
- Gower E. et al.. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 61, 45–57 (2014). - PubMed
-
- Wise M. et al.. Changing trends in hepatitis C–related mortality in the United States, 1995–2004. Hepatology 47, 1128–1135 (2008). - PubMed
-
- The Global Burden of Hepatitis C Working Group. The Global Burden of Hepatitis C. J Clinical Pharmacology 44, 20–29 (2004). - PubMed
-
- Afdhal N. et al.. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med 370, 1483–1493 (2014). - PubMed
-
- Afdhal N. et al.. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med 370, 1889–1898 (2014). - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

