Gene Regulation and Quality Control in Murine Polyomavirus Infection
- PMID: 27763514
- PMCID: PMC5086616
- DOI: 10.3390/v8100284
Gene Regulation and Quality Control in Murine Polyomavirus Infection
Abstract
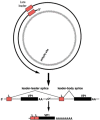
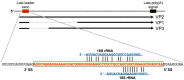
Murine polyomavirus (MPyV) infects mouse cells and is highly oncogenic in immunocompromised hosts and in other rodents. Its genome is a small, circular DNA molecule of just over 5000 base pairs and it encodes only seven polypeptides. While seemingly simply organized, this virus has adopted an unusual genome structure and some unusual uses of cellular quality control pathways that, together, allow an amazingly complex and varied pattern of gene regulation. In this review we discuss how MPyV leverages these various pathways to control its life cycle.
Keywords: RNA decay; RNA editing; nuclear retention; quality control; transcription.
Conflict of interest statement
The author declares no conflict of interest.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

