Drought-Responsive Mechanisms in Plant Leaves Revealed by Proteomics
- PMID: 27763546
- PMCID: PMC5085738
- DOI: 10.3390/ijms17101706
Drought-Responsive Mechanisms in Plant Leaves Revealed by Proteomics
Abstract
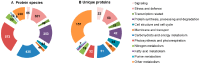
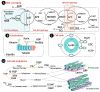
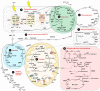
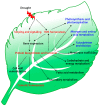
Plant drought tolerance is a complex trait that requires a global view to understand its underlying mechanism. The proteomic aspects of plant drought response have been extensively investigated in model plants, crops and wood plants. In this review, we summarize recent proteomic studies on drought response in leaves to reveal the common and specialized drought-responsive mechanisms in different plants. Although drought-responsive proteins exhibit various patterns depending on plant species, genotypes and stress intensity, proteomic analyses show that dominant changes occurred in sensing and signal transduction, reactive oxygen species scavenging, osmotic regulation, gene expression, protein synthesis/turnover, cell structure modulation, as well as carbohydrate and energy metabolism. In combination with physiological and molecular results, proteomic studies in leaves have helped to discover some potential proteins and/or metabolic pathways for drought tolerance. These findings provide new clues for understanding the molecular basis of plant drought tolerance.
Keywords: drought stress; leaves; molecular mechanism; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Farooq M., Wahid A., Kobayashi N., Fujita D., Basra S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009;29:185–212. doi: 10.1051/agro:2008021. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

