Structural Interface Forms and Their Involvement in Stabilization of Multidomain Proteins or Protein Complexes
- PMID: 27763556
- PMCID: PMC5085769
- DOI: 10.3390/ijms17101741
Structural Interface Forms and Their Involvement in Stabilization of Multidomain Proteins or Protein Complexes
Abstract
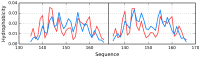
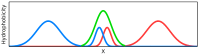
The presented analysis concerns the inter-domain and inter-protein interface in protein complexes. We propose extending the traditional understanding of the protein domain as a function of local compactness with an additional criterion which refers to the presence of a well-defined hydrophobic core. Interface areas in selected homodimers vary with respect to their contribution to share as well as individual (domain-specific) hydrophobic cores. The basic definition of a protein domain, i.e., a structural unit characterized by tighter packing than its immediate environment, is extended in order to acknowledge the role of a structured hydrophobic core, which includes the interface area. The hydrophobic properties of interfaces vary depending on the status of interacting domains-In this context we can distinguish: (1) Shared hydrophobic cores (spanning the whole dimer); (2) Individual hydrophobic cores present in each monomer irrespective of whether the dimer contains a shared core. Analysis of interfaces in dystrophin and utrophin indicates the presence of an additional quasi-domain with a prominent hydrophobic core, consisting of fragments contributed by both monomers. In addition, we have also attempted to determine the relationship between the type of interface (as categorized above) and the biological function of each complex. This analysis is entirely based on the fuzzy oil drop model.
Keywords: domain; dystrophin; homodimers; hydrophobic core; hydrophobicity; interface; utrophin.
Conflict of interest statement
Authors declare no conflict of interest.
Figures








References
-
- Levinthal C. Are there pathways for protein folding? J. Chem. Phys. 1968;65:44–45.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

