miR-125b modulates megakaryocyte maturation by targeting the cell-cycle inhibitor p19INK4D
- PMID: 27763644
- PMCID: PMC5133966
- DOI: 10.1038/cddis.2016.288
miR-125b modulates megakaryocyte maturation by targeting the cell-cycle inhibitor p19INK4D
Abstract
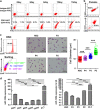
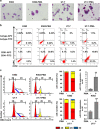
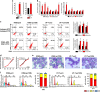
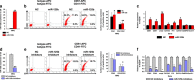
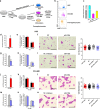
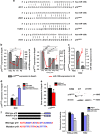
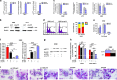
A better understanding of the mechanisms involved in megakaryocyte maturation will facilitate the generation of platelets in vitro and their clinical applications. A microRNA, miR-125b, has been suggested to have important roles in the self-renewal of megakaryocyte-erythroid progenitors and in platelet generation. However, miR-125b is also critical for hematopoietic stem cell self-renewal. Thus, the function of miR-125b and the complex signaling pathways regulating megakaryopoiesis remain to be elucidated. In this study, an attentive examination of the endogenous expression of miR-125b during megakaryocyte differentiation was performed. Accordingly, the differentiation of hematopoietic stem cells requires the downregulation of miR-125b, whereas megakaryocyte determination and maturation synchronize with miR-125b accumulation. The overexpression of miR-125b improves megakaryocytic differentiation of K562 and UT-7 cells. Furthermore, stage-specific overexpression of miR-125b in primary cells demonstrates that miR-125b mediates an enhancement of megakaryocytic differentiation after megakaryocyte determination, the stage at which megakaryocytes are negative for the expression of the hematopoietic progenitor marker CD34. The identification of miR-125b targets during megakaryopoiesis was focused on negative regulators of cell cycle because the transition of the G1/S phase has been associated with megakaryocyte polyploidization. Real-time PCR, western blot and luciferase reporter assay reveal that p19INK4D is a direct target of miR-125b. P19INK4D knockdown using small interfering RNA (siRNA) in megakaryocyte-induced K562 cells, UT-7 cells and CD61+ promegakaryocytes results in S-phase progression and increased polyploidy, as well as improved megakaryocyte differentiation, similarly to the effects of miR-125b overexpression. P19INK4D overexpression reverses these effects, as indicated by reduced expression of megakaryocyte markers, G1-phase arrest and polyploidy decrease. P19INK4D knockdown in miR-125b downregulated cells or p19INK4D overexpression in miR-125b upregulated cells rescued the effect of miR-125b. Taken together, these findings suggest that miR-125b expression positively regulates megakaryocyte development since the initial phases of megakaryocyte determination, and p19INK4D is one of the key mediators of miR-125b activity during the onset of megakaryocyte polyploidization.
Figures


Similar articles
-
miRNAs can increase the efficiency of ex vivo platelet generation.Ann Hematol. 2012 Nov;91(11):1673-84. doi: 10.1007/s00277-012-1517-z. Epub 2012 Jul 5. Ann Hematol. 2012. PMID: 22763947
-
p19 INK4d controls hematopoietic stem cells in a cell-autonomous manner during genotoxic stress and through the microenvironment during aging.Stem Cell Reports. 2014 Dec 9;3(6):1085-102. doi: 10.1016/j.stemcr.2014.10.005. Epub 2014 Nov 20. Stem Cell Reports. 2014. PMID: 25458892 Free PMC article.
-
P19INK4D links endomitotic arrest and megakaryocyte maturation and is regulated by AML-1.Blood. 2008 Apr 15;111(8):4081-91. doi: 10.1182/blood-2007-09-113266. Epub 2008 Feb 14. Blood. 2008. PMID: 18276842
-
Genesis of clone size heterogeneity in megakaryocytic and other hemopoietic colonies: the stochastic model revisited.Exp Hematol. 2001 Nov;29(11):1256-69. doi: 10.1016/s0301-472x(01)00728-7. Exp Hematol. 2001. PMID: 11698121 Review.
-
Intrinsic and extrinsic regulation of the proliferation and differentiation of cells in the rodent rostral migratory stream.J Neurosci Res. 2002 Sep 15;69(6):795-802. doi: 10.1002/jnr.10336. J Neurosci Res. 2002. PMID: 12205673 Free PMC article. Review.
Cited by
-
RHOB regulates megakaryocytic and erythroid differentiation by altering the cell cycle and cytoskeleton.Sci Rep. 2025 Apr 16;15(1):13159. doi: 10.1038/s41598-025-95946-3. Sci Rep. 2025. PMID: 40240762 Free PMC article.
-
Megakaryocyte ontogeny: Clinical and molecular significance.Exp Hematol. 2018 May;61:1-9. doi: 10.1016/j.exphem.2018.02.003. Epub 2018 Mar 2. Exp Hematol. 2018. PMID: 29501467 Free PMC article. Review.
-
CDKN1A as a potential target for Eltrombopag treatment in ITP and its regulation of the communication between macrophages and transitional B cells in ITP.Ann Hematol. 2025 Jun;104(6):3183-3197. doi: 10.1007/s00277-025-06436-5. Epub 2025 Jun 14. Ann Hematol. 2025. PMID: 40515824 Free PMC article.
-
The accumulation of miR-125b-5p is indispensable for efficient erythroblast enucleation.Cell Death Dis. 2022 Oct 21;13(10):886. doi: 10.1038/s41419-022-05331-5. Cell Death Dis. 2022. PMID: 36270980 Free PMC article.
-
miR-1915-3p regulates megakaryocytic and erythroid differentiation by targeting SOCS4.Thromb J. 2024 Aug 9;22(1):74. doi: 10.1186/s12959-024-00615-6. Thromb J. 2024. PMID: 39123189 Free PMC article.
References
-
- Bartel DP, Micrornas. Genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. - PubMed
-
- Havelange V, Garzon R. Micrornas: emerging key regulators of hematopoiesis. Am J Hematol 2010; 85: 935–942. - PubMed
-
- Emmrich S, Henke K, Hegermann J, Ochs M, Reinhardt D, Klusmann J-H. Mirnas can increase the efficiency of ex vivo platelet generation. Ann Hematol 2012; 91: 1673–1684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

