Application of Droplet Digital PCR for Estimating Vector Copy Number States in Stem Cell Gene Therapy
- PMID: 27763786
- PMCID: PMC5111482
- DOI: 10.1089/hgtb.2016.059
Application of Droplet Digital PCR for Estimating Vector Copy Number States in Stem Cell Gene Therapy
Abstract
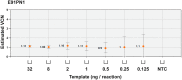
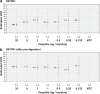
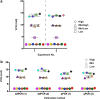
Stable gene transfer into target cell populations via integrating viral vectors is widely used in stem cell gene therapy (SCGT). Accurate vector copy number (VCN) estimation has become increasingly important. However, existing methods of estimation such as real-time quantitative PCR are more restricted in practicality, especially during clinical trials, given the limited availability of sample materials from patients. This study demonstrates the application of an emerging technology called droplet digital PCR (ddPCR) in estimating VCN states in the context of SCGT. Induced pluripotent stem cells (iPSCs) derived from a patient with X-linked chronic granulomatous disease were used as clonable target cells for transduction with alpharetroviral vectors harboring codon-optimized CYBB cDNA. Precise primer-probe design followed by multiplex analysis conferred assay specificity. Accurate estimation of per-cell VCN values was possible without reliance on a reference standard curve. Sensitivity was high and the dynamic range of detection was wide. Assay reliability was validated by observation of consistent, reproducible, and distinct VCN clustering patterns for clones of transduced iPSCs with varying numbers of transgene copies. Taken together, use of ddPCR appears to offer a practical and robust approach to VCN estimation with a wide range of clinical and research applications.
Keywords: chronic granulomatous disease; droplet digital PCR; induced pluripotent stem cells; integrating vectors; vector copy number.
Conflict of interest statement
Author Disclosure Y.Y. and S.I. are currently employed by Bio-Rad Laboratories.
Figures


References
-
- Cantsilieris S, Baird PN, White SJ. Molecular methods for genotyping complex copy number polymorphisms. Genomics 2013;101:86–93 - PubMed
-
- VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008;44:619–626 - PubMed
-
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 2008;3:1101–1108 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

