A Large Genome-Wide Association Study of Age-Related Hearing Impairment Using Electronic Health Records
- PMID: 27764096
- PMCID: PMC5072625
- DOI: 10.1371/journal.pgen.1006371
A Large Genome-Wide Association Study of Age-Related Hearing Impairment Using Electronic Health Records
Abstract
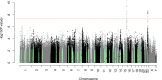
Age-related hearing impairment (ARHI), one of the most common sensory disorders, can be mitigated, but not cured or eliminated. To identify genetic influences underlying ARHI, we conducted a genome-wide association study of ARHI in 6,527 cases and 45,882 controls among the non-Hispanic whites from the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. We identified two novel genome-wide significant SNPs: rs4932196 (odds ratio = 1.185, p = 4.0x10-11), 52Kb 3' of ISG20, which replicated in a meta-analysis of the other GERA race/ethnicity groups (1,025 cases, 12,388 controls, p = 0.00094) and in a UK Biobank case-control analysis (30,802 self-reported cases, 78,586 controls, p = 0.015); and rs58389158 (odds ratio = 1.132, p = 1.8x10-9), which replicated in the UK Biobank (p = 0.00021). The latter SNP lies just outside exon 8 and is highly correlated (r2 = 0.96) with the missense SNP rs5756795 in exon 7 of TRIOBP, a gene previously associated with prelingual nonsyndromic hearing loss. We further tested these SNPs in phenotypes from audiologist notes available on a subset of GERA (4,903 individuals), stratified by case/control status, to construct an independent replication test, and found a significant effect of rs58389158 on speech reception threshold (SRT; overall GERA meta-analysis p = 1.9x10-6). We also tested variants within exons of 132 other previously-identified hearing loss genes, and identified two common additional significant SNPs: rs2877561 (synonymous change in ILDR1, p = 6.2x10-5), which replicated in the UK Biobank (p = 0.00057), and had a significant GERA SRT (p = 0.00019) and speech discrimination score (SDS; p = 0.0019); and rs9493627 (missense change in EYA4, p = 0.00011) which replicated in the UK Biobank (p = 0.0095), other GERA groups (p = 0.0080), and had a consistent significant result for SRT (p = 0.041) and suggestive result for SDS (p = 0.081). Large cohorts with GWAS data and electronic health records may be a useful method to characterize the genetic architecture of ARHI.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

