MIR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2
- PMID: 27764573
- PMCID: PMC5324854
- DOI: 10.1080/15548627.2016.1241922
MIR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2
Abstract
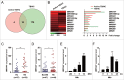
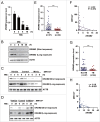
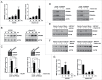
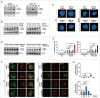
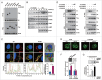
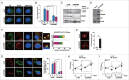
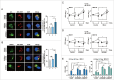
Autophagy is an important antimicrobial effector process that defends against Mycobacterium tuberculosis (Mtb), the human pathogen causing tuberculosis (TB). MicroRNAs (miRNAs), endogenous noncoding RNAs, are involved in various biological functions and act as post-transcriptional regulators to target mRNAs. The process by which miRNAs affect antibacterial autophagy and host defense mechanisms against Mtb infections in human monocytes and macrophages is largely uncharacterized. In this study, we show that Mtb significantly induces the expression of MIR144*/hsa-miR-144-5p, which targets the 3'-untranslated region of DRAM2 (DNA damage regulated autophagy modulator 2) in human monocytes and macrophages. Mtb infection downregulated, whereas the autophagy activators upregulated, DRAM2 expression in human monocytes and macrophages by activating AMP-activated protein kinase. In addition, overexpression of MIR144* decreased DRAM2 expression and formation of autophagosomes in human monocytes, whereas inhibition of MIR144* had the opposite effect. Moreover, the levels of MIR144* were elevated, whereas DRAM2 levels were reduced, in human peripheral blood cells and tissues in TB patients, indicating the clinical significance of MIR144* and DRAM2 in human TB. Notably, DRAM2 interacted with BECN1 and UVRAG, essential components of the autophagic machinery, leading to displacement of RUBCN from the BECN1 complex and enhancement of Ptdlns3K activity. Furthermore, MIR144* and DRAM2 were critically involved in phagosomal maturation and enhanced antimicrobial effects against Mtb. Our findings identify a previously unrecognized role of human MIR144* in the inhibition of antibacterial autophagy and the innate host immune response to Mtb. Additionally, these data reveal that DRAM2 is a key coordinator of autophagy activation that enhances antimicrobial activity against Mtb.
Keywords: AMPK; DRAM2; MIR144*; Mycobacterium tuberculosis; tuberculosis.
Figures

References
-
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 2011; 240:92-104; PMID:21349088; http://dx.doi.org/ 10.1111/j.1600-065X.2010.00995.x - DOI - PMC - PubMed
-
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; PMID:21248839; http://dx.doi.org/ 10.1038/nature09782 - DOI - PMC - PubMed
-
- Cemma M, Brumell JH. Interactions of pathogenic bacteria with autophagy systems. Curr Biol 2012; 22:R540-5; PMID:22790007; http://dx.doi.org/ 10.1016/j.cub.2012.06.001 - DOI - PubMed
-
- Deretic V. Autophagy, an immunologic magic bullet: Mycobacterium tuberculosis phagosome maturation block and how to bypass it. Future Microbiol 2008; 3:517-24; PMID:18811236; http://dx.doi.org/ 10.2217/17460913.3.5.517 - DOI - PMC - PubMed
-
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013; 501:512-6; PMID:24005326; http://dx.doi.org/ 10.1038/nature12566 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
