PDGFR-alpha inhibits melanoma growth via CXCL10/IP-10: a multi-omics approach
- PMID: 27764787
- PMCID: PMC5363585
- DOI: 10.18632/oncotarget.12629
PDGFR-alpha inhibits melanoma growth via CXCL10/IP-10: a multi-omics approach
Abstract
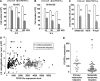
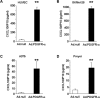
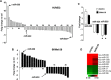
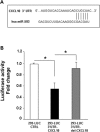
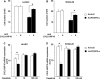
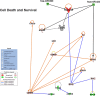
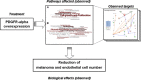
Melanoma is the most aggressive skin-cancer, showing high mortality at advanced stages. Platelet Derived Growth Factor Receptor-alpha (PDGFR-alpha) potently inhibits melanoma- and endothelium-proliferation and its expression is significantly reduced in melanoma-biopsies, suggesting that melanoma progression eliminates cells expressing PDGFR-alpha. In the present study transient overexpression of PDGFR-alpha in endothelial (HUVEC) and melanoma (SKMel-28, A375, Preyer) human-cells shows strong anti-proliferative effects, with profound transcriptome and miRNome deregulation. PDGFR-alpha overexpression strongly affects expression of 82 genes in HUVEC (41 up-, 41 down-regulated), and 52 genes in SKMel-28 (43 up-, 9 down-regulated). CXCL10/IP-10 transcript showed up to 20 fold-increase, with similar changes detectable at the protein level. miRNA expression profiling in cells overexpressing PDGFR-alpha identified 14 miRNAs up- and 40 down-regulated, with miR-503 being the most down-regulated (6.4 fold-reduction). miR-503, miR-630 and miR-424 deregulation was confirmed by qRT-PCR. Interestingly, the most upregulated transcript (i.e., CXCL10/IP-10) was a validated miR-503 target and CXCL10/IP-10 neutralization significantly reverted the anti-proliferative action of PDGFR-alpha, and PDGFR-alpha inhibition by Dasatinb totally reverted the CXCL10/IP10 induction, further supporting a functional interplay of these factors. Finally, integration of transcriptomics and miRNomics data highlighted several pathways affected by PDGFR-alpha.This study demonstrates for the first time that PDGFR-alpha strongly inhibits endothelial and melanoma cells proliferation in a CXCL10/IP-10 dependent way, via miR-503 down-regulation.
Keywords: angiogenesis; cancer; melanoma; miRNA; omics.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

