Attractor landscape analysis of colorectal tumorigenesis and its reversion
- PMID: 27765040
- PMCID: PMC5072344
- DOI: 10.1186/s12918-016-0341-9
Attractor landscape analysis of colorectal tumorigenesis and its reversion
Abstract
Background: Colorectal cancer arises from the accumulation of genetic mutations that induce dysfunction of intracellular signaling. However, the underlying mechanism of colorectal tumorigenesis driven by genetic mutations remains yet to be elucidated.
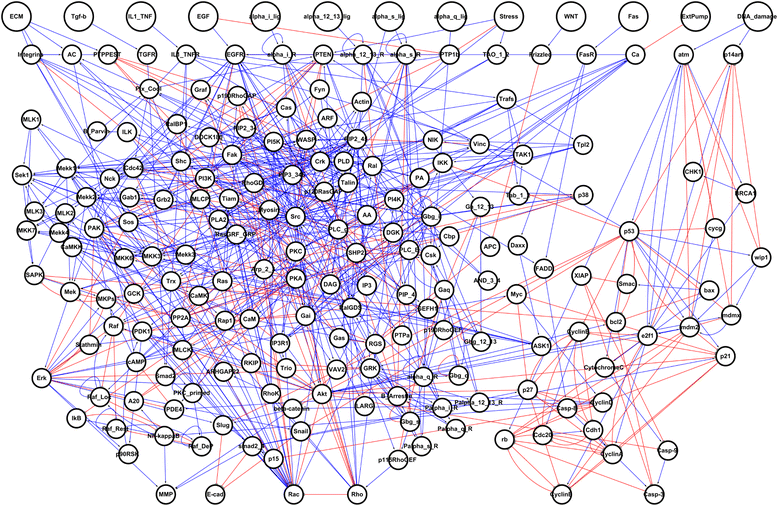
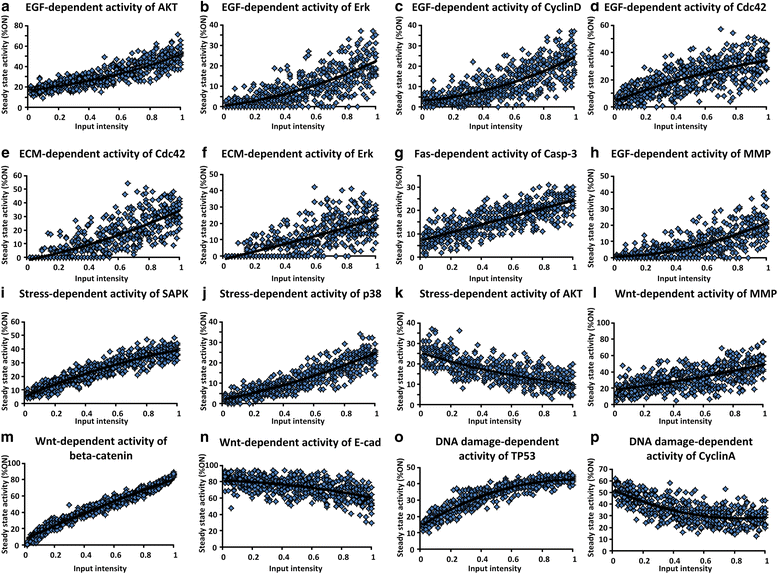
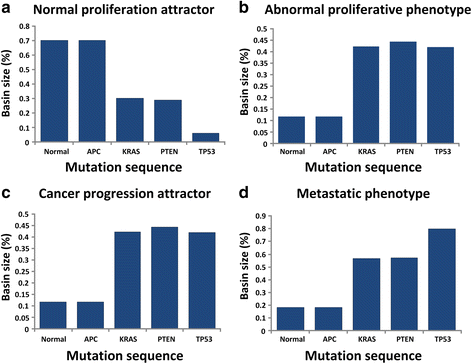
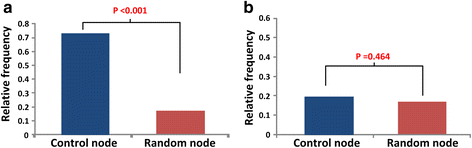
Results: To investigate colorectal tumorigenesis at a system-level, we have reconstructed a large-scale Boolean network model of the human signaling network by integrating previous experimental results on canonical signaling pathways related to proliferation, metastasis, and apoptosis. Throughout an extensive simulation analysis of the attractor landscape of the signaling network model, we found that the attractor landscape changes its shape by expanding the basin of attractors for abnormal proliferation and metastasis along with the accumulation of driver mutations. A further hypothetical study shows that restoration of a normal phenotype might be possible by reversely controlling the attractor landscape. Interestingly, the targets of approved anti-cancer drugs were highly enriched in the identified molecular targets for the reverse control.
Conclusions: Our results show that the dynamical analysis of a signaling network based on attractor landscape is useful in acquiring a system-level understanding of tumorigenesis and developing a new therapeutic strategy.
Keywords: Attractor landscape analysis; Cancer reversion; Colorectal tumorigenesis; Human signaling network; Reverse control; Systems biology.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

