Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression
- PMID: 27765066
- PMCID: PMC5073889
- DOI: 10.1186/s13059-016-1070-5
Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression
Erratum in
-
Erratum to: Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression.Genome Biol. 2016 Dec 1;17(1):249. doi: 10.1186/s13059-016-1113-y. Genome Biol. 2016. PMID: 27908289 Free PMC article. No abstract available.
Abstract
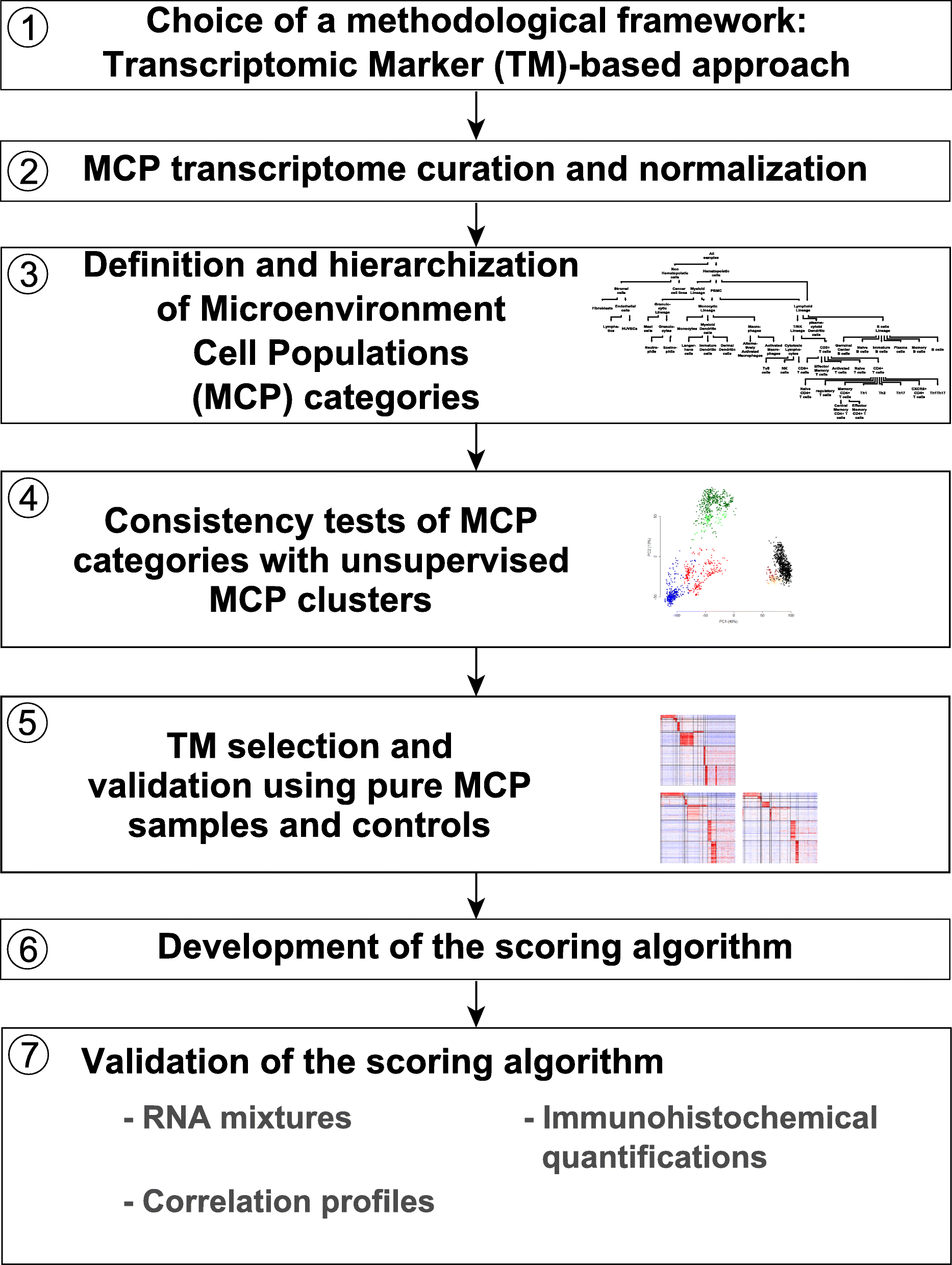
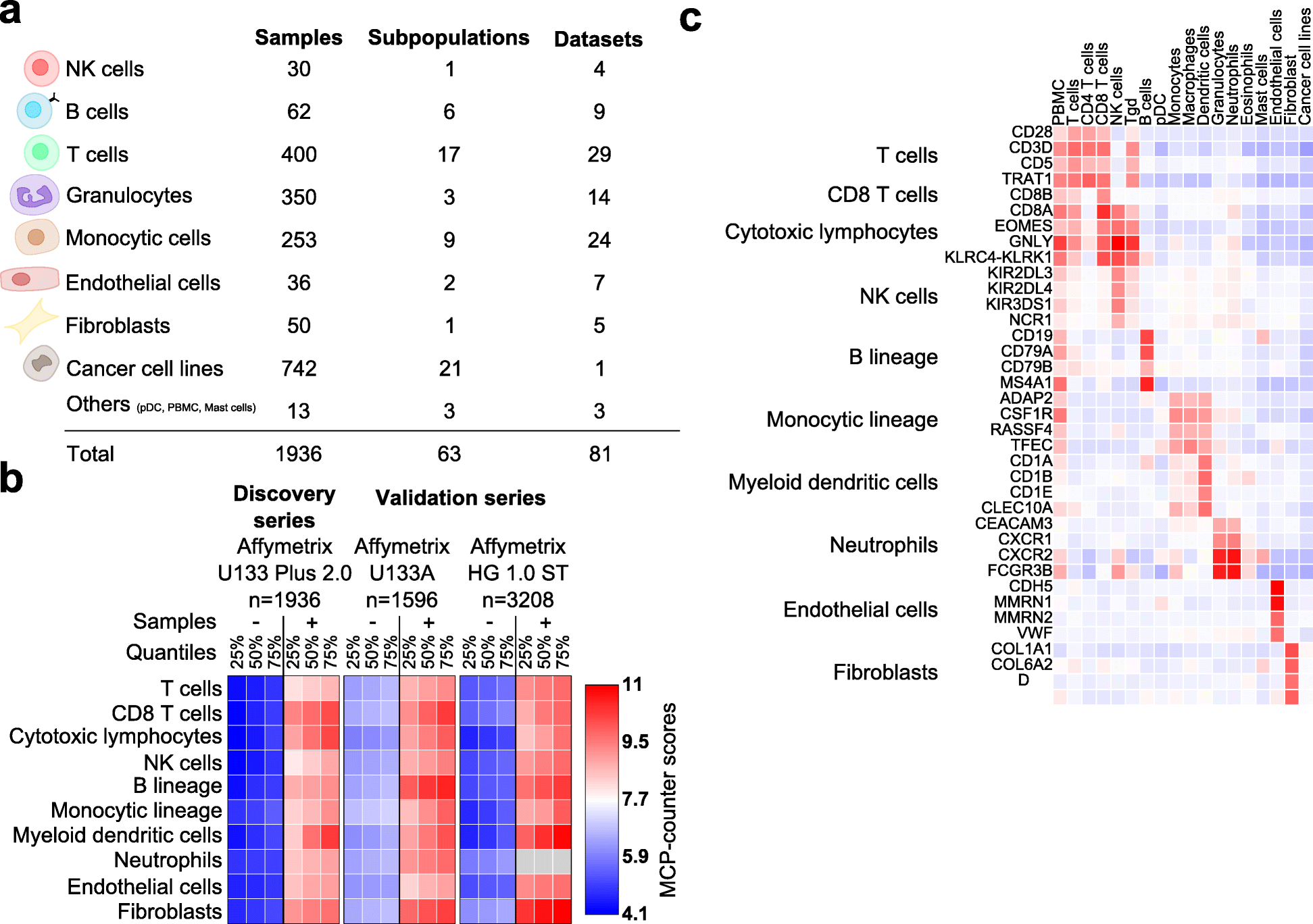
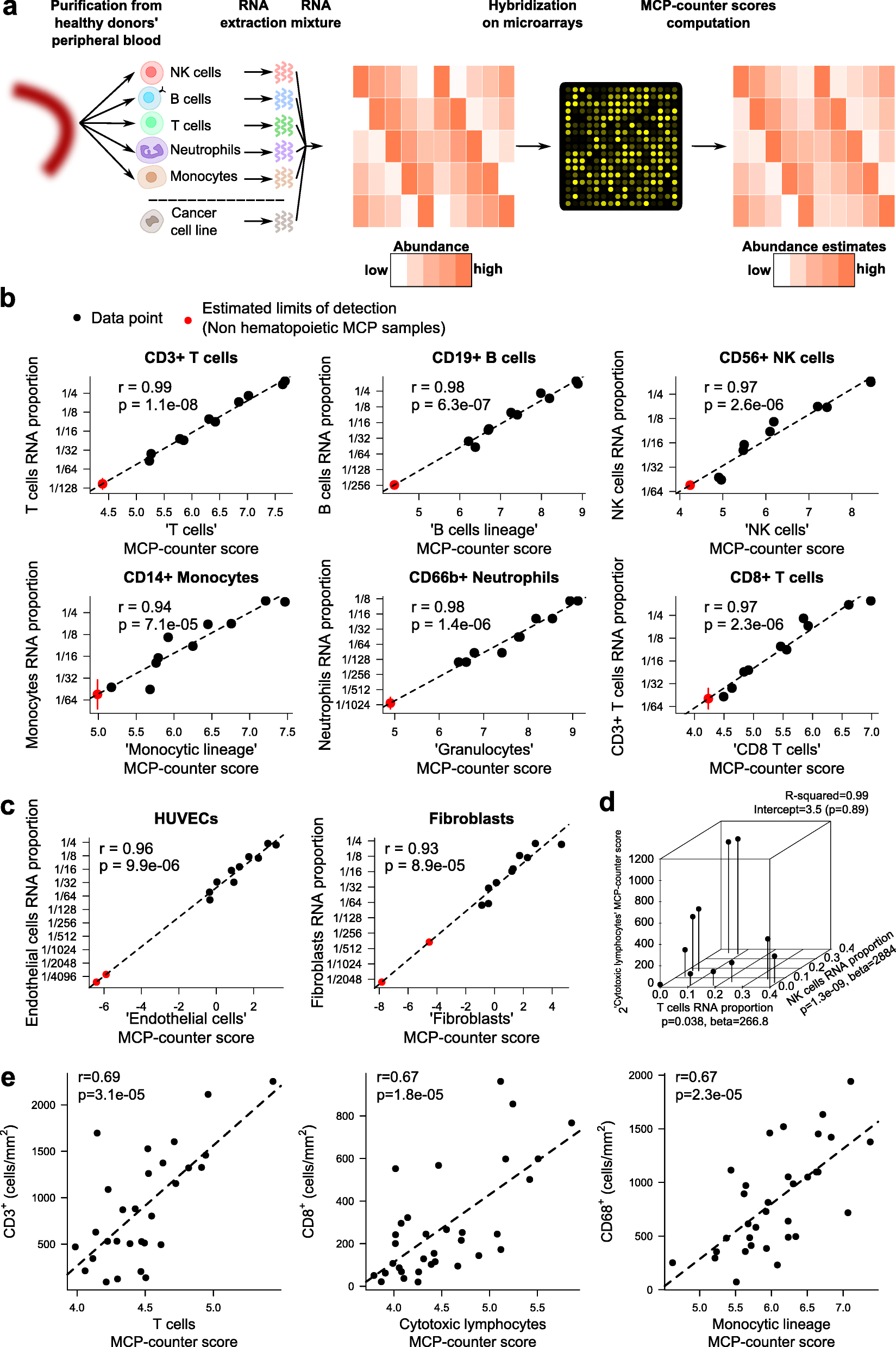
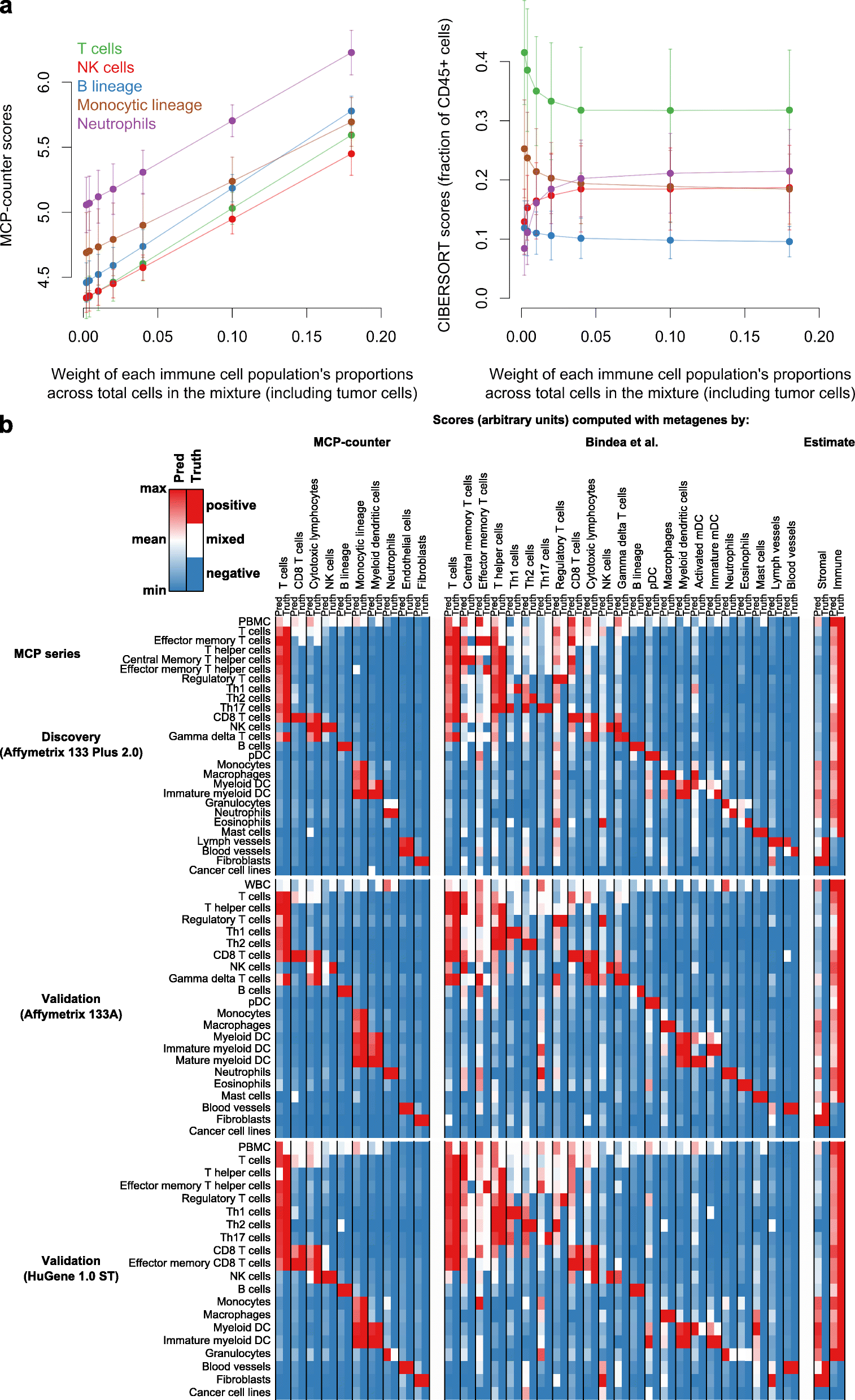
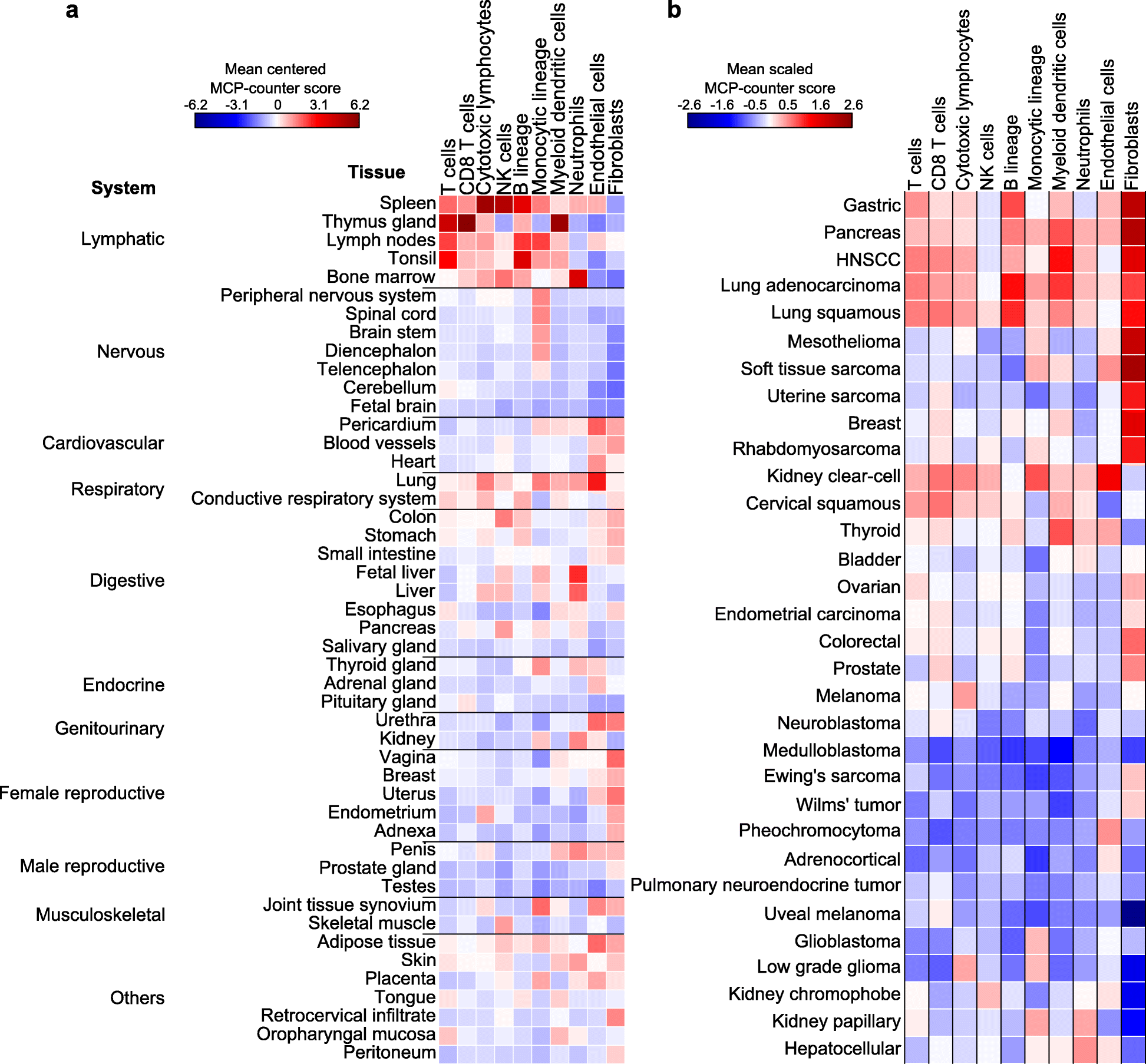
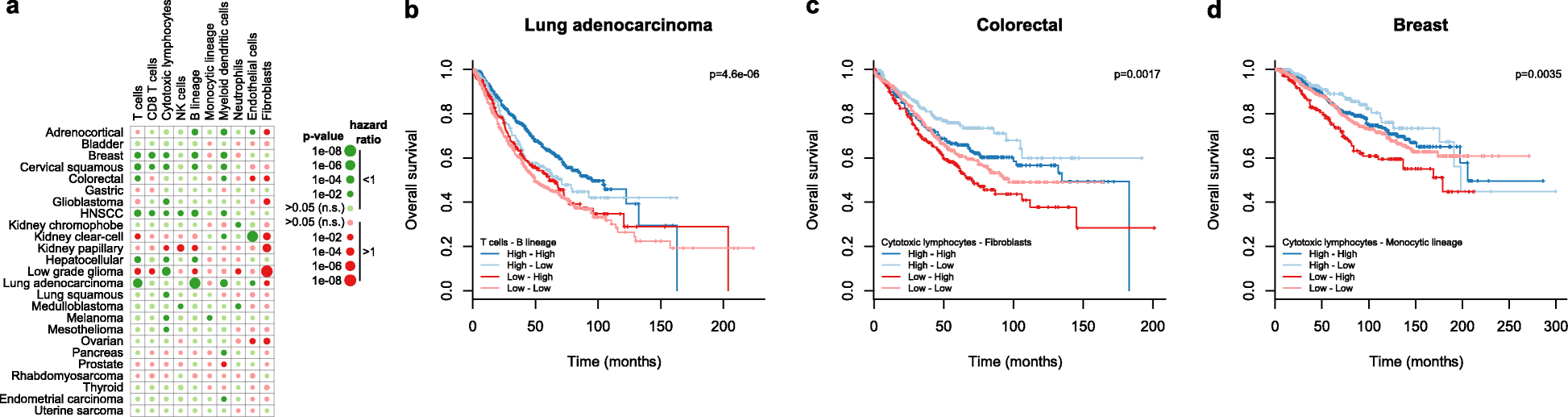
We introduce the Microenvironment Cell Populations-counter (MCP-counter) method, which allows the robust quantification of the absolute abundance of eight immune and two stromal cell populations in heterogeneous tissues from transcriptomic data. We present in vitro mRNA mixture and ex vivo immunohistochemical data that quantitatively support the validity of our method's estimates. Additionally, we demonstrate that MCP-counter overcomes several limitations or weaknesses of previously proposed computational approaches. MCP-counter is applied to draw a global picture of immune infiltrates across human healthy tissues and non-hematopoietic human tumors and recapitulates microenvironment-based patient stratifications associated with overall survival in lung adenocarcinoma and colorectal and breast cancer.
Keywords: Deconvolution; Gene signatures; Transcriptomic markers; Tumor microenvironment.
Figures
References
-
- Becht E, Giraldo NA, Germain C, de Reyniès A, Laurent-Puig P, Zucman-Rossi J, Dieu-Nosjean M, Sautès-Fridman C, Fridman WH. Immune contexture, immunoscore, and malignant cell molecular subgroups for prognostic and theranostic classifications of cancers. Adv Immunol. 2016;130:95–190. doi: 10.1016/bs.ai.2015.12.002. - DOI - PubMed
-
- Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, Meatchi T, Bruneval P, Cugnenc P, Trajanoski Z, Fridman W, Galon J. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–66. doi: 10.1056/NEJMoa051424. - DOI - PubMed
-
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoué F, Bruneval P, Cugnenc P, Trajanoski Z, Fridman W, Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. - DOI - PubMed
-
- Giraldo NA, Becht E, Pagès F, Skliris G, Verkarre V, Vano Y, Mejean A, Saint-Aubert N, Lacroix L, Natario I, Lupo A, Alifano M, Damotte D, Cazes A, Triebel F, Freeman GJ, Dieu-Nosjean M, Oudard S, Fridman WH, Sautès-Fridman C. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer. Clin Cancer Res. 2015;21:3031–40. doi: 10.1158/1078-0432.CCR-14-2926. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

