Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules
- PMID: 27766851
- PMCID: PMC5515585
- DOI: 10.1021/acs.jctc.6b00819
Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules
Abstract
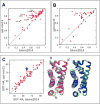
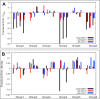
Most biomolecular modeling energy functions for structure prediction, sequence design, and molecular docking have been parametrized using existing macromolecular structural data; this contrasts molecular mechanics force fields which are largely optimized using small-molecule data. In this study, we describe an integrated method that enables optimization of a biomolecular modeling energy function simultaneously against small-molecule thermodynamic data and high-resolution macromolecular structural data. We use this approach to develop a next-generation Rosetta energy function that utilizes a new anisotropic implicit solvation model, and an improved electrostatics and Lennard-Jones model, illustrating how energy functions can be considerably improved in their ability to describe large-scale energy landscapes by incorporating both small-molecule and macromolecule data. The energy function improves performance in a wide range of protein structure prediction challenges, including monomeric structure prediction, protein-protein and protein-ligand docking, protein sequence design, and prediction of the free energy changes by mutation, while reasonably recapitulating small-molecule thermodynamic properties.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
