Evaluating the mobility potential of antibiotic resistance genes in environmental resistomes without metagenomics
- PMID: 27767072
- PMCID: PMC5073339
- DOI: 10.1038/srep35790
Evaluating the mobility potential of antibiotic resistance genes in environmental resistomes without metagenomics
Abstract
Antibiotic resistance genes are ubiquitous in the environment. However, only a fraction of them are mobile and able to spread to pathogenic bacteria. Until now, studying the mobility of antibiotic resistance genes in environmental resistomes has been challenging due to inadequate sensitivity and difficulties in contig assembly of metagenome based methods. We developed a new cost and labor efficient method based on Inverse PCR and long read sequencing for studying mobility potential of environmental resistance genes. We applied Inverse PCR on sediment samples and identified 79 different MGE clusters associated with the studied resistance genes, including novel mobile genetic elements, co-selected resistance genes and a new putative antibiotic resistance gene. The results show that the method can be used in antibiotic resistance early warning systems. In comparison to metagenomics, Inverse PCR was markedly more sensitive and provided more data on resistance gene mobility and co-selected resistances.
Figures
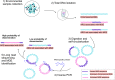



References
-
- World Health Organization. Antimicrobial resistance: global report on surveillance (WHO, 2014).
-
- D’Costa V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011). - PubMed
-
- Martinez J. L. Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

