Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system
- PMID: 27767081
- PMCID: PMC5073330
- DOI: 10.1038/srep35766
Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system
Abstract
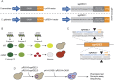
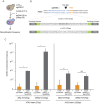
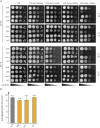
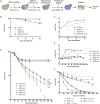
Among Candida species, the opportunistic fungal pathogen Candida glabrata has become the second most common causative agent of candidiasis in the world and a major public health concern. Yet, few molecular tools and resources are available to explore the biology of C. glabrata and to better understand its virulence during infection. In this study, we describe a robust experimental strategy to generate loss-of-function mutants in C. glabrata. The procedure is based on the development of three main tools: (i) a recombinant strain of C. glabrata constitutively expressing the CRISPR-Cas9 system, (ii) an online program facilitating the selection of the most efficient guide RNAs for a given C. glabrata gene, and (iii) the identification of mutant strains by the Surveyor technique and sequencing. As a proof-of-concept, we have tested the virulence of some mutants in vivo in a Drosophila melanogaster infection model. Our results suggest that yps11 and a previously uncharacterized serine/threonine kinase are involved, directly or indirectly, in the ability of the pathogenic yeast to infect this model host organism.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

