C9orf72 plays a central role in Rab GTPase-dependent regulation of autophagy
- PMID: 27768524
- PMCID: PMC5997165
- DOI: 10.1080/21541248.2016.1240495
C9orf72 plays a central role in Rab GTPase-dependent regulation of autophagy
Abstract
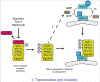
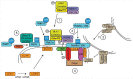
A GGGGCC hexanucleotide repeat expansion in the first intron of the C9orf72 gene is the most common genetic defect associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (C9ALS/FTD). Haploinsufficiency and a resulting loss of C9orf72 protein function has been suggested as a possible pathogenic mechanism in C9ALS/FTD. C9ALS/FTD patients exhibit specific ubiquitin and p62/sequestosome-1 positive but TDP-43 negative inclusions in the cerebellum and hippocampus, indicating possible autophagy deficits in these patients. In a recent study, we investigated this possibility by reducing expression of C9orf72 in cell lines and primary neurons and found that C9orf72 regulates the initiation of autophagy. C9orf72 interacts with Rab1a, preferentially in its GTP-bound state, as well as the ULK1 autophagy initiation complex. As an effector of Rab1a, C9orf72 controls the Rab1a-dependent trafficking of the ULK1 initiation complex prior to autophagosome formation. In line with this function, C9orf72 depletion in cell lines and primary neurons caused the accumulation of p62/sequestosome-1-positive inclusions. In support of a role in disease pathogenesis, C9ALS/FTD patient-derived iNeurons showed markedly reduced levels of autophagy. In this Commentary we summarise recent findings supporting the key role of C9orf72 in Rab GTPase-dependent regulation of autophagy and discuss autophagy dysregulation as a pathogenic mechanism in ALS/FTD.
Keywords: ALS; C9orf72; FTD; Rab GTPase; ULK1; autophagy.
Figures


References
-
- Swinnen B, Robberecht W. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol 2014; 10:661-70; PMID:25311585; https://doi.org/10.1038/nrneurol.2014.184 - DOI - PubMed
-
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-Linked FTD and ALS. Neuron 2011; 72:245-56; PMID:21944778; https://doi.org/10.1016/j.neuron.2011.09.011 - DOI - PMC - PubMed
-
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al.. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72:257-68; PMID:21944779; https://doi.org/10.1016/j.neuron.2011.09.010 - DOI - PMC - PubMed
-
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry Na, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al.. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 2013; 80:415-28; PMID:24139042; https://doi.org/10.1016/j.neuron.2013.10.015 - DOI - PMC - PubMed
-
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, et al.. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 2013; 110:E4530-9; PMID:24170860; https://doi.org/10.1073/pnas.1318835110 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
