Dom34 Links Translation to Protein O-mannosylation
- PMID: 27768707
- PMCID: PMC5074521
- DOI: 10.1371/journal.pgen.1006395
Dom34 Links Translation to Protein O-mannosylation
Abstract
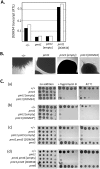
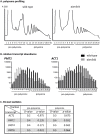
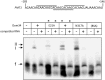
In eukaryotes, Dom34 upregulates translation by securing levels of activatable ribosomal subunits. We found that in the yeast Saccharomyces cerevisiae and the human fungal pathogen Candida albicans, Dom34 interacts genetically with Pmt1, a major isoform of protein O-mannosyltransferase. In C. albicans, lack of Dom34 exacerbated defective phenotypes of pmt1 mutants, while they were ameliorated by Dom34 overproduction that enhanced Pmt1 protein but not PMT1 transcript levels. Translational effects of Dom34 required the 5'-UTR of the PMT1 transcript, which bound recombinant Dom34 directly at a CA/AC-rich sequence and regulated in vitro translation. Polysomal profiling revealed that Dom34 stimulates general translation moderately, but that it is especially required for translation of transcripts encoding Pmt isoforms 1, 4 and 6. Because defective protein N- or O-glycosylation upregulates transcription of PMT genes, it appears that Dom34-mediated specific translational upregulation of the PMT transcripts optimizes cellular responses to glycostress. Its translational function as an RNA binding protein acting at the 5'-UTR of specific transcripts adds another facet to the known ribosome-releasing functions of Dom34 at the 3'-UTR of transcripts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

