IBMPFD Disease-Causing Mutant VCP/p97 Proteins Are Targets of Autophagic-Lysosomal Degradation
- PMID: 27768726
- PMCID: PMC5074563
- DOI: 10.1371/journal.pone.0164864
IBMPFD Disease-Causing Mutant VCP/p97 Proteins Are Targets of Autophagic-Lysosomal Degradation
Erratum in
-
Correction: IBMPFD Disease-Causing Mutant VCP/p97 Proteins Are Targets of Autophagic-Lysosomal Degradation.PLoS One. 2016 Nov 18;11(11):e0167192. doi: 10.1371/journal.pone.0167192. eCollection 2016. PLoS One. 2016. PMID: 27861584 Free PMC article.
Abstract
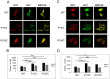
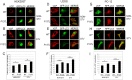
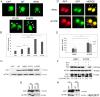
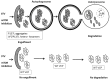
The ubiquitin-proteasome system (UPS) degrades soluble proteins and small aggregates, whereas macroautophagy (autophagy herein) eliminates larger protein aggregates, tangles and even whole organelles in a lysosome-dependent manner. VCP/p97 was implicated in both pathways. VCP/p97 mutations cause a rare multisystem disease called IBMPFD (Inclusion Body Myopathy with Paget's Disease and Frontotemporal Dementia). Here, we studied the role IBMPFD-related mutants of VCP/p97 in autophagy. In contrast with the wild-type VCP/p97 protein or R155C or R191Q mutants, the P137L mutant was aggregate-prone. We showed that, unlike commonly studied R155C or R191Q mutants, the P137L mutant protein stimulated both autophagosome and autolysosome formation. Moreover, P137L mutant protein itself was a substrate of autophagy. Starvation- and mTOR inhibition-induced autophagy led to the degradation of the P137L mutant protein, while preserving the wild-type and functional VCP/p97. Strikingly, similar to the P137L mutant, other IBMPFD-related VCP/p97 mutants, namely R93C and G157R mutants induced autophagosome and autolysosome formation; and G157R mutant formed aggregates that could be cleared by autophagy. Therefore, cellular phenotypes caused by P137L mutant expression were not isolated observations, and some other IBMPFD disease-related VCP/p97 mutations could lead to similar outcomes. Our results indicate that cellular mechanisms leading to IBMPFD disease may be various, and underline the importance of studying different disease-associated mutations in order to better understand human pathologies and tailor mutation-specific treatment strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Wang Q, Song C, Li CC. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol. 2004;146(1–2):44–57. Epub 2004/03/24. [pii]. . - PubMed
-
- Watts GD, Thorne M, Kovach MJ, Pestronk A, Kimonis VE. Clinical and genetic heterogeneity in chromosome 9p associated hereditary inclusion body myopathy: exclusion of GNE and three other candidate genes. Neuromuscul Disord. 2003;13(7–8):559–67. Epub 2003/08/19. S0960896603000701 [pii]. . - PubMed
-
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36(4):377–81. Epub 2004/03/23. [pii]. . - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

