Progressive Loss of Function in a Limb Enhancer during Snake Evolution
- PMID: 27768887
- PMCID: PMC5484524
- DOI: 10.1016/j.cell.2016.09.028
Progressive Loss of Function in a Limb Enhancer during Snake Evolution
Abstract
The evolution of body shape is thought to be tightly coupled to changes in regulatory sequences, but specific molecular events associated with major morphological transitions in vertebrates have remained elusive. We identified snake-specific sequence changes within an otherwise highly conserved long-range limb enhancer of Sonic hedgehog (Shh). Transgenic mouse reporter assays revealed that the in vivo activity pattern of the enhancer is conserved across a wide range of vertebrates, including fish, but not in snakes. Genomic substitution of the mouse enhancer with its human or fish ortholog results in normal limb development. In contrast, replacement with snake orthologs caused severe limb reduction. Synthetic restoration of a single transcription factor binding site lost in the snake lineage reinstated full in vivo function to the snake enhancer. Our results demonstrate changes in a regulatory sequence associated with a major body plan transition and highlight the role of enhancers in morphological evolution. PAPERCLIP.
Keywords: CRISPR/Cas9; Sonic hedgehog (Shh); ZRS; cis-regulatory element; enhancer; evo-devo; genome editing; limb development; morphological evolution; snakes.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


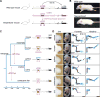
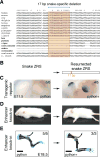

Comment in
-
Unwinding Limb Development.Cell. 2016 Oct 20;167(3):598-600. doi: 10.1016/j.cell.2016.10.007. Cell. 2016. PMID: 27768881
-
Evolution: Enhanced Footing for Snake Limb Development.Curr Biol. 2016 Dec 5;26(23):R1237-R1240. doi: 10.1016/j.cub.2016.10.018. Curr Biol. 2016. PMID: 27923134
References
-
- Acemel RD, Tena JJ, Irastorza-Azcarate I, Marlétaz F, Gomez-Marin C, la Calle-Mustienes de E, Bertrand S, Diaz SG, Aldea D, Aury J-M, et al. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet. 2016;48:336–341. - PubMed
-
- Apesteguía S, Zaher H. A Cretaceous terrestrial snake with robust hindlimbs and a sacrum. Nature. 2006;440:1037–1040. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

