Biofuel metabolic engineering with biosensors
- PMID: 27768949
- PMCID: PMC5161612
- DOI: 10.1016/j.cbpa.2016.09.020
Biofuel metabolic engineering with biosensors
Abstract
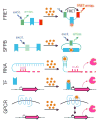
Metabolic engineering offers the potential to renewably produce important classes of chemicals, particularly biofuels, at an industrial scale. DNA synthesis and editing techniques can generate large pathway libraries, yet identifying the best variants is slow and cumbersome. Traditionally, analytical methods like chromatography and mass spectrometry have been used to evaluate pathway variants, but such techniques cannot be performed with high throughput. Biosensors - genetically encoded components that actuate a cellular output in response to a change in metabolite concentration - are therefore a promising tool for rapid and high-throughput evaluation of candidate pathway variants. Applying biosensors can also dynamically tune pathways in response to metabolic changes, improving balance and productivity. Here, we describe the major classes of biosensors and briefly highlight recent progress in applying them to biofuel-related metabolic pathway engineering.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures


References
-
- Keasling JD. Manufacturing Molecules Through Metabolic Engineering. Science. 2010;330:1355–1358. - PubMed
-
- Liu R, Bassalo MC, Zeitoun RI, Gill RT. Genome scale engineering techniques for metabolic engineering. Metab Eng. 2015;32:143–154. - PubMed
-
- Hounslow E, Noirel J, Gilmour DJ, Wright PC. Lipid quantification techniques for screening oleaginous species of microalgae for biofuel production. Eur J Lipid Sci Technol. 2016 doi: 10.1002/ejlt.201500469. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

