Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation
- PMID: 27770024
- PMCID: PMC5207059
- DOI: 10.1074/jbc.M116.755256
Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation
Abstract
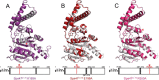
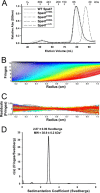
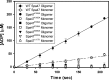
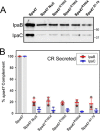
Like many Gram-negative pathogens, Shigella rely on a complex type III secretion system (T3SS) to inject effector proteins into host cells, take over host functions, and ultimately establish infection. Despite these critical roles, the energetics and regulatory mechanisms controlling the T3SS and pathogen virulence remain largely unclear. In this study, we present a series of high resolution crystal structures of Spa47 and use the structures to model an activated Spa47 oligomer, finding that ATP hydrolysis may be supported by specific side chain contributions from adjacent protomers within the complex. Follow-up mutagenesis experiments targeting the predicted active site residues validate the oligomeric model and determined that each of the tested residues are essential for Spa47 ATPase activity, although they are not directly responsible for stable oligomer formation. Although N-terminal domain truncation was necessary for crystal formation, it resulted in strictly monomeric Spa47 that is unable to hydrolyze ATP, despite maintaining the canonical ATPase core structure and active site residues. Coupled with studies of ATPase inactive full-length Spa47 point mutants, we find that Spa47 oligomerization and ATP hydrolysis are needed for complete T3SS apparatus formation, a proper translocator secretion profile, and Shigella virulence. This work represents the first structure-function characterization of Spa47, uniquely complementing the multitude of included Shigella T3SS phenotype assays and providing a more complete understanding of T3SS ATPase-mediated pathogen virulence. Additionally, these findings provide a strong platform for follow-up studies evaluating regulation of Spa47 oligomerization in vivo as a much needed means of treating and perhaps preventing shigellosis.
Keywords: ATPase; Shigella; Spa47; arginine finger; crystal structure; oligomerization; secretion regulation; type III secretion apparatus (T3SA); type III secretion system (T3SS).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Portaliou A. G., Tsolis K. C., Loos M. S., Zorzini V., and Economou A. (2016) Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 41, 175–189 - PubMed
-
- Cossart P., and Sansonetti P. J. (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248 - PubMed
-
- World Health Organization (Updated April 2013) Diarrhoeal Disease, Fact Sheet no. 330, http://www.who.int/mediacentre/factsheets/fs330/en/
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

