A computational interactome and functional annotation for the human proteome
- PMID: 27770567
- PMCID: PMC5115866
- DOI: 10.7554/eLife.18715
A computational interactome and functional annotation for the human proteome
Abstract
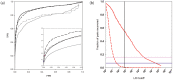
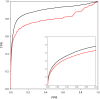
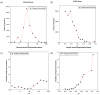
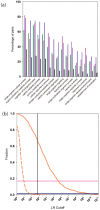
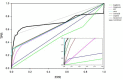
We present a database, PrePPI (Predicting Protein-Protein Interactions), of more than 1.35 million predicted protein-protein interactions (PPIs). Of these at least 127,000 are expected to constitute direct physical interactions although the actual number may be much larger (~500,000). The current PrePPI, which contains predicted interactions for about 85% of the human proteome, is related to an earlier version but is based on additional sources of interaction evidence and is far larger in scope. The use of structural relationships allows PrePPI to infer numerous previously unreported interactions. PrePPI has been subjected to a series of validation tests including reproducing known interactions, recapitulating multi-protein complexes, analysis of disease associated SNPs, and identifying functional relationships between interacting proteins. We show, using Gene Set Enrichment Analysis (GSEA), that predicted interaction partners can be used to annotate a protein's function. We provide annotations for most human proteins, including many annotated as having unknown function.
Keywords: computational biology; function annotation; human; machine learning; protein interactions; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

References
-
- Altenhoff AM, Škunca N, Glover N, Train CM, Sueki A, Piližota I, Gori K, Tomiczek B, Müller S, Redestig H, Gonnet GH, Dessimoz C. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Research. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. - DOI - PMC - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Beigi F, Schmeckpeper J, Pow-Anpongkul P, Payne JA, Zhang L, Zhang Z, Huang J, Mirotsou M, Dzau VJ. C3orf58, a novel paracrine protein, stimulates cardiomyocyte cell-cycle progression through the PI3K-AKT-CDK7 pathway. Circulation Research. 2013;113:372–380. doi: 10.1161/CIRCRESAHA.113.301075. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

