Contralateral migration of oculomotor neurons is regulated by Slit/Robo signaling
- PMID: 27770832
- PMCID: PMC5075191
- DOI: 10.1186/s13064-016-0073-y
Contralateral migration of oculomotor neurons is regulated by Slit/Robo signaling
Abstract
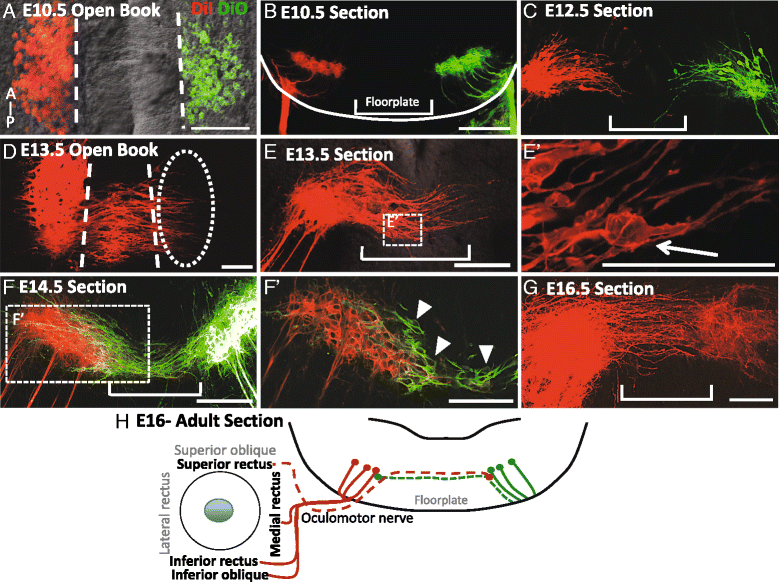
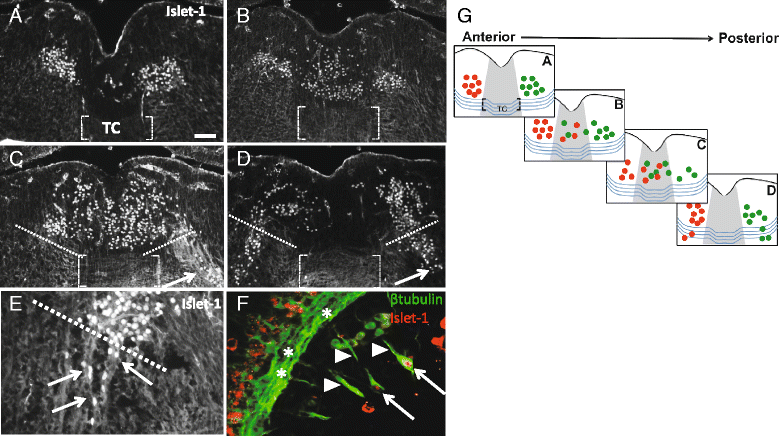
Background: Oculomotor neurons develop initially like typical motor neurons, projecting axons out of the ventral midbrain to their ipsilateral targets, the extraocular muscles. However, in all vertebrates, after the oculomotor nerve (nIII) has reached the extraocular muscle primordia, the cell bodies that innervate the superior rectus migrate to join the contralateral nucleus. This motor neuron migration represents a unique strategy to form a contralateral motor projection. Whether migration is guided by diffusible cues remains unknown.
Methods: We examined the role of Slit chemorepellent signals in contralateral oculomotor migration by analyzing mutant mouse embryos.
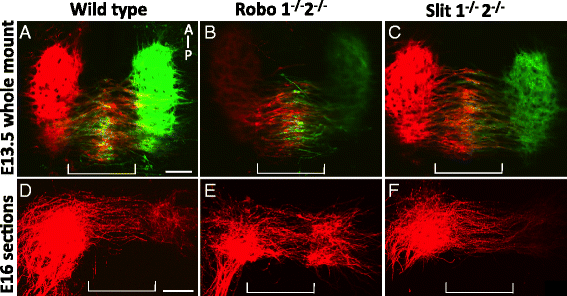
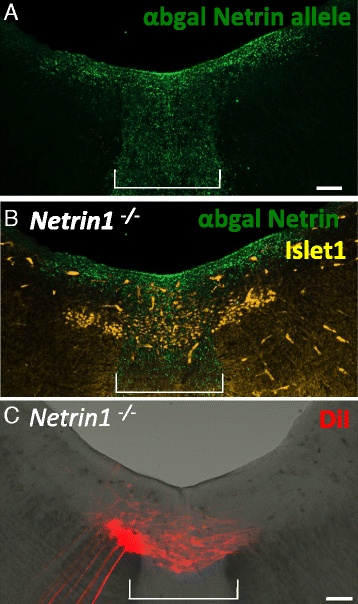
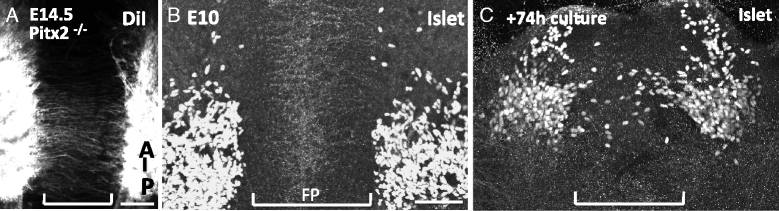
Results: We found that the ventral midbrain expresses high levels of both Slit1 and 2, and that oculomotor neurons express the repellent Slit receptors Robo1 and Robo2. Therefore, Slit signals are in a position to influence the migration of oculomotor neurons. In Slit 1/2 or Robo1/2 double mutant embryos, motor neuron cell bodies migrated into the ventral midbrain on E10.5, three days prior to normal migration. These early migrating neurons had leading projections into and across the floor plate. In contrast to the double mutants, embryos which were mutant for single Slit or Robo genes did not have premature migration or outgrowth on E10.5, demonstrating a cooperative requirement of Slit1 and 2, as well as Robo1 and 2. To test how Slit/Robo midline repulsion is modulated, we found that the normal migration did not require the receptors Robo3 and CXCR4, or the chemoattractant, Netrin 1. The signal to initiate contralateral migration is likely autonomous to the midbrain because oculomotor neurons migrate in embryos that lack either nerve outgrowth or extraocular muscles, or in cultured midbrains that lacked peripheral tissue.
Conclusion: Overall, our results demonstrate that a migratory subset of motor neurons respond to floor plate-derived Slit repulsion to properly control the timing of contralateral migration.
Keywords: Floor plate; Migration; Motor neuron; Oculomotor; Slit/Robo.
Figures






References
-
- Hogg ID. Observations of the development of the nucleus of Edinger-Westphal in man and the albino rat. J Comp Neurol. 1966;126:567–584. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

