Reaction kinetic analysis of the 3-hydroxypropionate/4-hydroxybutyrate CO2 fixation cycle in extremely thermoacidophilic archaea
- PMID: 27771364
- PMCID: PMC5433351
- DOI: 10.1016/j.ymben.2016.10.009
Reaction kinetic analysis of the 3-hydroxypropionate/4-hydroxybutyrate CO2 fixation cycle in extremely thermoacidophilic archaea
Abstract
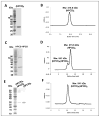
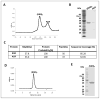
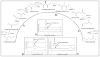
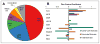
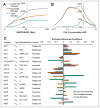
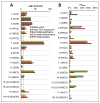
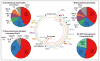
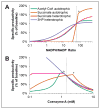
The 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) cycle fixes CO2 in extremely thermoacidophilic archaea and holds promise for metabolic engineering because of its thermostability and potentially rapid pathway kinetics. A reaction kinetics model was developed to examine the biological and biotechnological attributes of the 3HP/4HB cycle as it operates in Metallosphaera sedula, based on previous information as well as on kinetic parameters determined here for recombinant versions of five of the cycle enzymes (malonyl-CoA/succinyl-CoA reductase, 3-hydroxypropionyl-CoA synthetase, 3-hydroxypropionyl-CoA dehydratase, acryloyl-CoA reductase, and succinic semialdehyde reductase). The model correctly predicted previously observed features of the cycle: the 35-65% split of carbon flux through the acetyl-CoA and succinate branches, the high abundance and relative ratio of acetyl-CoA/propionyl-CoA carboxylase (ACC) and MCR, and the significance of ACC and hydroxybutyryl-CoA synthetase (HBCS) as regulated control points for the cycle. The model was then used to assess metabolic engineering strategies for incorporating CO2 into chemical intermediates and products of biotechnological importance: acetyl-CoA, succinate, and 3-hydroxypropionate.
Keywords: 3-hydroxypropionate; 4-hydroxybutyrate; CO(2) fixation; Metallosphaera sedula.
Copyright © 2016 International Metabolic Engineering Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Amend JP, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev. 2001;25:175–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

