Expanding the archaellum regulatory network - the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius
- PMID: 27771939
- PMCID: PMC5300886
- DOI: 10.1002/mbo3.414
Expanding the archaellum regulatory network - the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius
Abstract
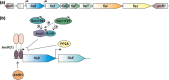
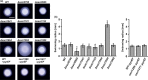
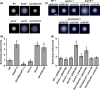
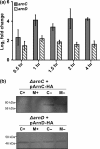
Expression of the archaellum, the archaeal-type IV pilus-like rotating motility structure is upregulated under nutrient limitation. This is controlled by a network of regulators, called the archaellum regulatory network (arn). Several of the components of this network in Sulfolobus acidocaldarius can be phosphorylated, and the deletion of the phosphatase PP2A results in strongly increased motility during starvation, indicating a role for phosphorylation in the regulation of motility. Analysis of the motility of different protein kinase deletion strains revealed that deletion of saci_0965, saci_1181, and saci_1193 resulted in reduced motility, whereas the deletion of saci_1694 resulted in hypermotility. Here ArnC (Saci_1193) and ArnD (Saci_1694) are characterized. Purified ArnC and ArnD phosphorylate serine and threonine residues in the C-terminus of the repressor ArnB. arnC is upregulated in starvation medium, whereas arnD is constitutively expressed. However, while differences in the expression and levels of flaB were observed in the ΔarnD strain during growth under rich conditions, under nutrient limiting conditions the ΔarnC and ΔarnD strains showed no large differences in the expression levels of the archaellum or of the studied regulators. This suggests that next to the regulation via the archaellum regulatory network additional regulatory mechanisms of expression and/or activity of the archaellum exist.
Keywords: S. acidocaldarius; archaeal flagella; archaellum regulation; phosphorylation; protein kinases; signaling network.
© 2016 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures


References
-
- Briza, P. , Bogengruber, E. , Thu, A. , Ru, M. , Mu, M. , & Breitenbach, M. (2002). Systematic analysis of sporulation phenotypes in 624 non‐lethal homozygous deletion strains of Saccharomyces cerevisiae . Yeast, 19, 403–422. - PubMed
-
- Brock, T. D. , Brock, K. M. , Belly, R. T. , & Weiss, R. L. (1972). Sulfolobus: A new genus of sulfur‐oxidizing bacteria living at low pH and high temperature. Archiv für Mikrobiologie, 84, 54–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

