Marked Differences in C9orf72 Methylation Status and Isoform Expression between C9/ALS Human Embryonic and Induced Pluripotent Stem Cells
- PMID: 27773700
- PMCID: PMC5106522
- DOI: 10.1016/j.stemcr.2016.09.011
Marked Differences in C9orf72 Methylation Status and Isoform Expression between C9/ALS Human Embryonic and Induced Pluripotent Stem Cells
Abstract
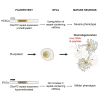
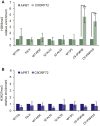
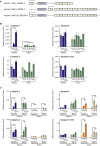
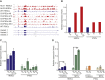
We established two human embryonic stem cell (hESC) lines with a GGGGCC expansion in the C9orf72 gene (C9), and compared them with haploidentical and unrelated C9 induced pluripotent stem cells (iPSCs). We found a marked difference in C9 methylation between the cells. hESCs and parental fibroblasts are entirely unmethylated while the iPSCs are hypermethylated. In addition, we show that the expansion alters promoter usage and interferes with the proper splicing of intron 1, eventually leading to the accumulation of repeat-containing mRNA following neural differentiation. These changes are attenuated in C9 iPSCs, presumably owing to hypermethylation. Altogether, this study highlights the importance of neural differentiation in the pathogenesis of disease and points to the potential role of hypermethylation as a neuroprotective mechanism against pathogenic mRNAs, envisaging a milder phenotype in C9 iPSCs.
Keywords: C9/ALS; CpG islands; DNA methylation; disease modelling; neurodegeneration; pluripotent stem cells; unstable repeat expansions.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
A C9ORF72 BAC mouse model recapitulates key epigenetic perturbations of ALS/FTD.Mol Neurodegener. 2017 Jun 12;12(1):46. doi: 10.1186/s13024-017-0185-9. Mol Neurodegener. 2017. PMID: 28606110 Free PMC article.
-
Neural Differentiation and spinal cord organoid generation from induced pluripotent stem cells (iPSCs) for ALS modelling and inflammatory screening.Mol Neurobiol. 2024 Jul;61(7):4732-4749. doi: 10.1007/s12035-023-03836-4. Epub 2023 Dec 21. Mol Neurobiol. 2024. PMID: 38127186
-
Generation of induced pluripotent stem cells (CSSi017-A)(12862) from an ALS patient carrying a repeat expansion in the C9orf72 gene.Stem Cell Res. 2024 Jun;77:103412. doi: 10.1016/j.scr.2024.103412. Epub 2024 Apr 10. Stem Cell Res. 2024. PMID: 38613988
-
Current progress and potential practical application for human pluripotent stem cells.Int Rev Cell Mol Biol. 2011;292:153-96. doi: 10.1016/B978-0-12-386033-0.00004-9. Int Rev Cell Mol Biol. 2011. PMID: 22078961 Review.
-
C9orf72 isoforms in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration.Brain Res. 2016 Sep 15;1647:43-49. doi: 10.1016/j.brainres.2016.04.062. Epub 2016 Apr 29. Brain Res. 2016. PMID: 27134035 Review.
Cited by
-
Epigenetic Changes in Prion and Prion-like Neurodegenerative Diseases: Recent Advances, Potential as Biomarkers, and Future Perspectives.Int J Mol Sci. 2022 Oct 20;23(20):12609. doi: 10.3390/ijms232012609. Int J Mol Sci. 2022. PMID: 36293477 Free PMC article. Review.
-
Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair.Hum Mol Genet. 2020 Aug 3;29(13):2200-2217. doi: 10.1093/hmg/ddaa106. Hum Mol Genet. 2020. PMID: 32504093 Free PMC article.
-
Representing Diversity in the Dish: Using Patient-Derived in Vitro Models to Recreate the Heterogeneity of Neurological Disease.Front Neurosci. 2018 Feb 9;12:56. doi: 10.3389/fnins.2018.00056. eCollection 2018. Front Neurosci. 2018. PMID: 29479303 Free PMC article. Review.
-
Characterization of C9orf72 haplotypes to evaluate the effects of normal and pathological variations on its expression and splicing.PLoS Genet. 2021 Mar 29;17(3):e1009445. doi: 10.1371/journal.pgen.1009445. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33780440 Free PMC article.
-
A C9ORF72 BAC mouse model recapitulates key epigenetic perturbations of ALS/FTD.Mol Neurodegener. 2017 Jun 12;12(1):46. doi: 10.1186/s13024-017-0185-9. Mol Neurodegener. 2017. PMID: 28606110 Free PMC article.
References
-
- Almeida S., Gascon E., Tran H., Chou H.J., Gendron T.F., Degroot S., Tapper A.R., Sellier C., Charlet-Berguerand N., Karydas A. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. - PMC - PubMed
-
- Bauer P.O. Methylation of C9orf72 expansion reduces RNA foci formation and dipeptide-repeat proteins expression in cells. Neurosci. Lett. 2016;612:204–209. - PubMed
-
- Belzil V.V., Bauer P.O., Prudencio M., Gendron T.F., Stetler C.T., Yan I.K., Pregent L., Daughrity L., Baker M.C., Rademakers R. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

