Fhit and Wwox loss-associated genome instability: A genome caretaker one-two punch
- PMID: 27773744
- PMCID: PMC7213024
- DOI: 10.1016/j.jbior.2016.09.008
Fhit and Wwox loss-associated genome instability: A genome caretaker one-two punch
Abstract
Expression of Fhit and Wwox protein is frequently lost or reduced in many human cancers. In this report, we provide data that further characterizes the molecular consequences of Fhit loss in the initiation of DNA double-strand breaks (DSBs), and of Wwox loss in altered repair of DSBs. We show that loss of Fhit initiates mild genome instability in early passage mouse kidney cells, confirming that DNA damage associated with Fhit-deficiency is not limited to cancer cells. We also demonstrate that the cause of Fhit-deficient DSBs: thymidine deficiency-induced replication stress, can be resolved with thymidine supplementation in early passage mouse kidney cells before extensive genome instability occurs. As for consequences of Wwox loss in cancer, we show in a small panel of breast cancer cells and mouse embryonic fibroblasts that Wwox expression predicts response to radiation and mitomycin C, all agents that cause DSBs. In addition, loss of Wwox significantly reduced progression free survival in a cohort of ovarian cancer patients treated with platin-based chemotherapies. Finally, stratification of a cohort of squamous lung cancers by Fhit expression reveals that Wwox expression is significantly reduced in the low Fhit-expressing group, suggesting that loss of Fhit is quickly succeeded by loss of Wwox. We propose that Fhit and Wwox loss work synergistically in cancer progression and that DNA damage caused by Fhit could be targeted early in cancer initiation for prevention, while DNA damage caused by Wwox loss could be targeted later in cancer progression, particularly in cancers that develop resistance to genotoxic therapies.
Keywords: Chromosome fragile site; DNA double-strand breaks; Fhit; Genome caretaker; Wwox.
Copyright © 2017. Published by Elsevier Ltd.
Figures



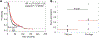
References
-
- Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM, 2004b. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 64, 8256–8261. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

