The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins
- PMID: 27773769
- PMCID: PMC5124403
- DOI: 10.1016/j.virusres.2016.10.010
The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins
Abstract
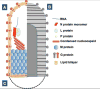
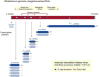
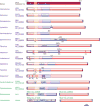
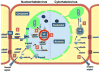
The family Rhabdoviridae consists of mostly enveloped, bullet-shaped or bacilliform viruses with a negative-sense, single-stranded RNA genome that infect vertebrates, invertebrates or plants. This ecological diversity is reflected by the diversity and complexity of their genomes. Five canonical structural protein genes are conserved in all rhabdoviruses, but may be overprinted, overlapped or interspersed with several novel and diverse accessory genes. This review gives an overview of the characteristics and diversity of rhabdoviruses, their taxonomic classification, replication mechanism, properties of classical rhabdoviruses such as rabies virus and rhabdoviruses with complex genomes, rhabdoviruses infecting aquatic species, and plant rhabdoviruses with both mono- and bipartite genomes.
Keywords: Diversity; Genome organization; Negative-sense RNA virus; Replication; Rhabdovirus; Taxonomy.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures

References
-
- Ahne W, Bjorklund H, Essbauer S, Fijan N, Kurath G, Winton J. Spring viremia of carp – review. Dis Aquat Organ. 2002;52:261–272. - PubMed
-
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

