C9orf72's Interaction with Rab GTPases-Modulation of Membrane Traffic and Autophagy
- PMID: 27774051
- PMCID: PMC5053994
- DOI: 10.3389/fncel.2016.00228
C9orf72's Interaction with Rab GTPases-Modulation of Membrane Traffic and Autophagy
Abstract
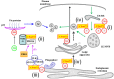
Hexanucleotide repeat expansion in an intron of Chromosome 9 open reading frame 72 (C9orf72) is the most common genetic cause of Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Dementia (FTD). While functional haploinsufficiency of C9orf72 resulting from the mutation may play a role in ALS/FTD, the actual cellular role of the protein has been unclear. Recent findings have now shown that C9orf72 physically and functionally interacts with multiple members of the Rab small GTPases family, consequently exerting important influences on cellular membrane traffic and the process of autophagy. Loss of C9orf72 impairs endocytosis in neuronal cell lines, and attenuated autophagosome formation. Interestingly, C9orf72 could influence autophagy both as part of a Guanine nucleotide exchange factor (GEF) complex, or as a Rab effector that facilitates transport of the Unc-51-like Autophagy Activating Kinase 1 (Ulk1) autophagy initiation complex. The cellular function of C9orf72 is discussed in the light of these recent findings.
Keywords: ALS; C9orf72; Rab; autophagy; membrane trafficking.
Figures
Similar articles
-
C9orf72, a protein associated with amyotrophic lateral sclerosis (ALS) is a guanine nucleotide exchange factor.PeerJ. 2018 Oct 17;6:e5815. doi: 10.7717/peerj.5815. eCollection 2018. PeerJ. 2018. PMID: 30356970 Free PMC article.
-
C9orf72 plays a central role in Rab GTPase-dependent regulation of autophagy.Small GTPases. 2018 Sep 3;9(5):399-408. doi: 10.1080/21541248.2016.1240495. Epub 2016 Oct 21. Small GTPases. 2018. PMID: 27768524 Free PMC article. Review.
-
The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy.EMBO J. 2016 Aug 1;35(15):1656-76. doi: 10.15252/embj.201694401. Epub 2016 Jun 22. EMBO J. 2016. PMID: 27334615 Free PMC article.
-
C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking.Hum Mol Genet. 2014 Jul 1;23(13):3579-95. doi: 10.1093/hmg/ddu068. Epub 2014 Feb 18. Hum Mol Genet. 2014. PMID: 24549040 Free PMC article.
-
Autophagy and neurodegeneration: Unraveling the role of C9ORF72 in the regulation of autophagy and its relationship to ALS-FTD pathology.Front Cell Neurosci. 2023 Mar 16;17:1086895. doi: 10.3389/fncel.2023.1086895. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37006471 Free PMC article. Review.
Cited by
-
Therapeutic Targeting of Rab GTPases: Relevance for Alzheimer's Disease.Biomedicines. 2022 May 16;10(5):1141. doi: 10.3390/biomedicines10051141. Biomedicines. 2022. PMID: 35625878 Free PMC article. Review.
-
Rab GTPases and Membrane Trafficking in Neurodegeneration.Curr Biol. 2018 Apr 23;28(8):R471-R486. doi: 10.1016/j.cub.2018.02.010. Curr Biol. 2018. PMID: 29689231 Free PMC article. Review.
-
Microglia and C9orf72 in neuroinflammation and ALS and frontotemporal dementia.J Clin Invest. 2017 Sep 1;127(9):3250-3258. doi: 10.1172/JCI90607. Epub 2017 Jul 24. J Clin Invest. 2017. PMID: 28737506 Free PMC article. Review.
-
Synaptopathy: presynaptic convergence in frontotemporal dementia and amyotrophic lateral sclerosis.Brain. 2024 Jul 5;147(7):2289-2307. doi: 10.1093/brain/awae074. Brain. 2024. PMID: 38451707 Free PMC article. Review.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

