Open PHACTS computational protocols for in silico target validation of cellular phenotypic screens: knowing the knowns
- PMID: 27774140
- PMCID: PMC5063042
- DOI: 10.1039/c6md00065g
Open PHACTS computational protocols for in silico target validation of cellular phenotypic screens: knowing the knowns
Abstract
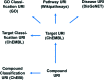
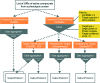
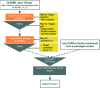
Phenotypic screening is in a renaissance phase and is expected by many academic and industry leaders to accelerate the discovery of new drugs for new biology. Given that phenotypic screening is per definition target agnostic, the emphasis of in silico and in vitro follow-up work is on the exploration of possible molecular mechanisms and efficacy targets underlying the biological processes interrogated by the phenotypic screening experiments. Herein, we present six exemplar computational protocols for the interpretation of cellular phenotypic screens based on the integration of compound, target, pathway, and disease data established by the IMI Open PHACTS project. The protocols annotate phenotypic hit lists and allow follow-up experiments and mechanistic conclusions. The annotations included are from ChEMBL, ChEBI, GO, WikiPathways and DisGeNET. Also provided are protocols which select from the IUPHAR/BPS Guide to PHARMACOLOGY interaction file selective compounds to probe potential targets and a correlation robot which systematically aims to identify an overlap of active compounds in both the phenotypic as well as any kinase assay. The protocols are applied to a phenotypic pre-lamin A/C splicing assay selected from the ChEMBL database to illustrate the process. The computational protocols make use of the Open PHACTS API and data and are built within the Pipeline Pilot and KNIME workflow tools.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

