Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts
- PMID: 27774290
- PMCID: PMC5014487
- DOI: 10.1093/ve/vev021
Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts
Abstract
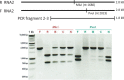
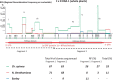
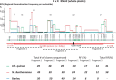
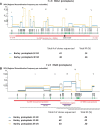
We have previously reported intra-segmental crossovers in Brome mosaic virus (BMV) RNAs. In this work, we studied the homologous recombination of BMV RNA in three different hosts: barley (Hordeum vulgare), Chenopodium quinoa, and Nicotiana benthamiana that were co-infected with two strains of BMV: Russian (R) and Fescue (F). Our work aimed at (1) establishing the frequency of recombination, (2) mapping the recombination hot spots, and (3) addressing host effects. The F and R nucleotide sequences differ from each other at many translationally silent nucleotide substitutions. We exploited this natural variability to track the crossover sites. Sequencing of a large number of cDNA clones revealed multiple homologous crossovers in each BMV RNA segment, in both the whole plants and protoplasts. Some recombination hot spots mapped at similar locations in different hosts, suggesting a role for viral factors, but other sites depended on the host. Our results demonstrate the chimeric ('mosaic') nature of the BMV RNA genome.
Keywords: Brome mosaic bromovirus; RNA replication; homologous RNA recombination; host effects; recombination frequency; recombination hot spots.
Figures

Similar articles
-
Homologous crossovers among molecules of brome mosaic bromovirus RNA1 or RNA2 segments in vivo.J Virol. 2005 May;79(9):5732-42. doi: 10.1128/JVI.79.9.5732-5742.2005. J Virol. 2005. PMID: 15827188 Free PMC article.
-
Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots.Virology. 1999 Feb 1;254(1):92-104. doi: 10.1006/viro.1998.9545. Virology. 1999. PMID: 9927577
-
Silencing homologous RNA recombination hot spots with GC-rich sequences in brome mosaic virus.J Virol. 1998 Feb;72(2):1122-30. doi: 10.1128/JVI.72.2.1122-1130.1998. J Virol. 1998. PMID: 9445008 Free PMC article.
-
RNA recombination in brome mosaic virus, a model plus strand RNA virus.Acta Biochim Pol. 1998;45(4):847-68. Acta Biochim Pol. 1998. PMID: 10397334 Review.
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
Cited by
-
Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs.Pathogens. 2022 Jul 21;11(7):817. doi: 10.3390/pathogens11070817. Pathogens. 2022. PMID: 35890061 Free PMC article.
-
Intermolecular RNA Recombination Occurs at Different Frequencies in Alternate Forms of Brome Mosaic Virus RNA Replication Compartments.Viruses. 2018 Mar 15;10(3):131. doi: 10.3390/v10030131. Viruses. 2018. PMID: 29543718 Free PMC article.
-
Recombination in Enteroviruses, a Multi-Step Modular Evolutionary Process.Viruses. 2019 Sep 14;11(9):859. doi: 10.3390/v11090859. Viruses. 2019. PMID: 31540135 Free PMC article. Review.
-
An Improved Brome mosaic virus Silencing Vector: Greater Insert Stability and More Extensive VIGS.Plant Physiol. 2018 Jan;176(1):496-510. doi: 10.1104/pp.17.00905. Epub 2017 Nov 10. Plant Physiol. 2018. PMID: 29127260 Free PMC article.
-
Characterization of Variant RNAs Encapsidated during Bromovirus Infection by High-Throughput Sequencing.Pathogens. 2024 Jan 22;13(1):96. doi: 10.3390/pathogens13010096. Pathogens. 2024. PMID: 38276169 Free PMC article.
References
-
- Awadalla P. (2003) ‘The Evolutionary Genomics of Pathogen Recombination’, Nature Reviews Genetics, 4: 50–60. - PubMed
-
- Bedhomme S., Hillung J., Elena S. F. (2015) ‘Emerging Viruses: Why They are Not Jacks of All Trades?’, Current Opinion in Virology, 10: 1–6. - PubMed
-
- Bujarski J. J. (1998) Bromovirus isolation. In: Foster G., Taylor S. (eds.) Plant Virology Protocols: From Virus Isolation to Transgenic Resistance, pp. 183-188. Totowa, NJ: Humana Press.
LinkOut - more resources
Full Text Sources
Other Literature Sources

