Evolution in spatially mixed host environments increases divergence for evolved fitness and intrapopulation genetic diversity in RNA viruses
- PMID: 27774292
- PMCID: PMC4989875
- DOI: 10.1093/ve/vev022
Evolution in spatially mixed host environments increases divergence for evolved fitness and intrapopulation genetic diversity in RNA viruses
Abstract
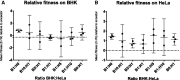
Virus populations may be challenged to evolve in spatially heterogeneous environments, such as mixtures of host cells that pose differing selection pressures. Spatial heterogeneity may select for evolved polymorphisms, where multiple virus subpopulations coexist by specializing on a narrow subset of the available hosts. Alternatively, spatial heterogeneity may select for evolved generalism, where a single genotype dominates the virus population by occupying a relatively broader host niche. In addition, the extent of spatial heterogeneity should influence the degree of divergence among virus populations encountering identical environmental challenges. Spatial heterogeneity creates environmental complexity that should increase the probability of differing adaptive phenotypic solutions, thus producing greater divergence among replicate virus populations, relative to counterparts evolving in strictly homogeneous host environments. Here, we tested these ideas using experimental evolution of RNA virus populations grown in laboratory tissue culture. We allowed vesicular stomatitis virus (VSV) lineages to evolve in replicated environments containing BHK-21 (baby hamster kidney) cells, HeLa (human epithelial) cells, or spatially heterogeneous host cell mixtures. Results showed that generalist phenotypes dominated in evolved virus populations across all treatments. Also, we observed greater variance in host-use performance (fitness) among VSV lineages evolved under spatial heterogeneity, relative to lineages evolved in homogeneous environments. Despite measurable differences in fitness, consensus Sanger sequencing revealed no fixed genetic differences separating the evolved lineages from their common ancestor. In contrast, deep sequencing of evolved VSV populations confirmed that the degree of divergence among replicate lineages was correlated with a larger number of minority variants. This correlation between divergence and the number of minority variants was significant only when we considered variants with a frequency of at least 10 per cent in the population. The number of lower-frequency minority variants per population did not significantly correlate with divergence.
Keywords: Vesicular stomatitis virus; adaptation; environmental heterogeneity; experimental evolution; niche breadth; quasispecies.
Figures
Similar articles
-
Gauging genetic diversity of generalists: A test of genetic and ecological generalism with RNA virus experimental evolution.Virus Evol. 2019 Jun 29;5(1):vez019. doi: 10.1093/ve/vez019. eCollection 2019 Jan. Virus Evol. 2019. PMID: 31275611 Free PMC article.
-
Generalized selection to overcome innate immunity selects for host breadth in an RNA virus.Evolution. 2016 Feb;70(2):270-81. doi: 10.1111/evo.12845. Epub 2016 Jan 20. Evolution. 2016. PMID: 26882316
-
Cost of host radiation in an RNA virus.Genetics. 2000 Dec;156(4):1465-70. doi: 10.1093/genetics/156.4.1465. Genetics. 2000. PMID: 11102349 Free PMC article.
-
Local adaptation of plant viruses: lessons from experimental evolution.Mol Ecol. 2017 Apr;26(7):1711-1719. doi: 10.1111/mec.13836. Epub 2016 Sep 22. Mol Ecol. 2017. PMID: 27612225 Review.
-
The emergence of performance trade-offs during local adaptation: insights from experimental evolution.Mol Ecol. 2017 Apr;26(7):1720-1733. doi: 10.1111/mec.13979. Epub 2017 Jan 13. Mol Ecol. 2017. PMID: 28029196 Review.
Cited by
-
Gauging genetic diversity of generalists: A test of genetic and ecological generalism with RNA virus experimental evolution.Virus Evol. 2019 Jun 29;5(1):vez019. doi: 10.1093/ve/vez019. eCollection 2019 Jan. Virus Evol. 2019. PMID: 31275611 Free PMC article.
-
Prior evolution in stochastic versus constant temperatures affects RNA virus evolvability at a thermal extreme.Ecol Evol. 2020 Apr 29;10(12):5440-5450. doi: 10.1002/ece3.6287. eCollection 2020 Jun. Ecol Evol. 2020. PMID: 32607165 Free PMC article.
-
Cooperative Virus-Virus Interactions: An Evolutionary Perspective.Biodes Res. 2022 Aug 9;2022:9819272. doi: 10.34133/2022/9819272. eCollection 2022. Biodes Res. 2022. PMID: 37850129 Free PMC article. Review.
-
Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change.Evolution. 2017 Apr;71(4):872-883. doi: 10.1111/evo.13193. Epub 2017 Feb 14. Evolution. 2017. PMID: 28121018 Free PMC article.
-
Culicoides-Specific Fitness Increase of Vesicular Stomatitis Virus in Insect-to-Insect Infections.Insects. 2024 Jan 5;15(1):34. doi: 10.3390/insects15010034. Insects. 2024. PMID: 38249040 Free PMC article.
References
-
- Agudelo-Romero P., de la Iglesia F., Elena S. F. (2008) ‘The Pleiotropic Cost of Host-Specialization in Tobacco etch potyvirus’, Infection, Genetics, and Evolution, 8: 806–14. - PubMed
-
- Alto B. W., et al. (2013) ‘Stochastic Temperatures Impede RNA Virus Adaptation’, Evolution, 67: 969–79. - PubMed
-
- Andrews S. (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data <http://www.bioinformatics.babraham.ac.uk/projects/fastqc/2010>.
-
- Bennet A. F., Lenski R. E., Mittler J. E. (1992) ‘Evolutionary Adaptation to Temperature. 1. Fitness Responses of Escherichia coli to Changes in its Thermal Environment’, Evolution, 46: 16–30. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

