Genetic diversity of STLV-2 and interspecies transmission of STLV-3 in wild-living bonobos
- PMID: 27774304
- PMCID: PMC4900509
- DOI: 10.1093/ve/vew011
Genetic diversity of STLV-2 and interspecies transmission of STLV-3 in wild-living bonobos
Abstract
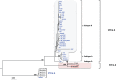
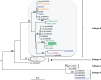
There are currently four known primate T-cell lymphotropic virus groups (PTLV1-4), each of which comprises closely related simian (STLV) and human (HTLV) viruses. For PTLV-1 and PTLV-3, simian and human viruses are interspersed, suggesting multiple cross-species transmission events; however, for PTLV-2 this is not so clear because HTLV-2 and STLV-2 strains from captive bonobos (Pan paniscus) form two distinct clades. To determine to what extent bonobos are naturally infected with STLV, we screened fecal samples (n = 633) from wild-living bonobos (n = 312) at six different sites in the Democratic Republic of Congo (DRC) for the presence of STLV nucleic acids. STLV infection was detected in 8 of 312 bonobos at four of six field sites, suggesting an overall prevalence of 2.6% (ranging from 0 to 8%). Six samples contained STLV-2, while the two others contained STLV-3, as determined by phylogenetic analysis of partial tax and Long Terminal Repeats (LTR) sequences. The new STLV-2 sequences were highly diverse, but grouped with previously identified STLV-2 strains as a sister clade to HTLV-2. In contrast, the new STLV-3 sequences did not cluster together, but were more closely related to STLVs from sympatric monkey species. These results show for the first time that fecal samples can be used to detect STLV infection in apes. These results also show that wild-living bonobos are endemically infected with STLV-2, but have acquired STLV-3 on at least two occasions most likely by cross-species transmission from monkey species on which they prey. Future studies of bonobos and other non-human primate species in Central Africa are needed to identify the simian precursor of HTLV-2 in humans.
Keywords: DRC; STLV-2; STLV-3; bonobos; feces.
Figures


References
-
- Alcantara L. C., et al. (2003) ‘Brazilian HTLV Type 2a Strains From Intravenous Drug Users (IDUs) Appear to Have Originated From Two Sources: Brazilian Amerindians and European/North American IDUs’, AIDS Research and Human Retroviruses, 19: 519–23. - PubMed
-
- Calvignac-Spencer S., et al. (2013) ‘Carrion Fly-Derived DNA as a Tool for Comprehensive and Cost-Effective Assessment of Mammalian Biodiversity’, Molecular Ecology, 22: 915–24. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

