Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system
- PMID: 27774985
- PMCID: PMC5079060
- DOI: 10.1038/ncomms13219
Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system
Abstract
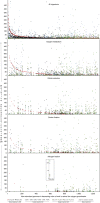
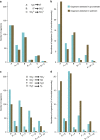
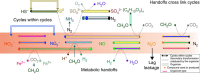
The subterranean world hosts up to one-fifth of all biomass, including microbial communities that drive transformations central to Earth's biogeochemical cycles. However, little is known about how complex microbial communities in such environments are structured, and how inter-organism interactions shape ecosystem function. Here we apply terabase-scale cultivation-independent metagenomics to aquifer sediments and groundwater, and reconstruct 2,540 draft-quality, near-complete and complete strain-resolved genomes that represent the majority of known bacterial phyla as well as 47 newly discovered phylum-level lineages. Metabolic analyses spanning this vast phylogenetic diversity and representing up to 36% of organisms detected in the system are used to document the distribution of pathways in coexisting organisms. Consistent with prior findings indicating metabolic handoffs in simple consortia, we find that few organisms within the community can conduct multiple sequential redox transformations. As environmental conditions change, different assemblages of organisms are selected for, altering linkages among the major biogeochemical cycles.
Figures



References
-
- McMahon S. & Parnell J. Weighing the deep continental biosphere. FEMS Microbiol. Ecol. 87, 113–120 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

