Single-Stranded Nucleic Acids Bind to the Tetramer Interface of SAMHD1 and Prevent Formation of the Catalytic Homotetramer
- PMID: 27775344
- PMCID: PMC5531264
- DOI: 10.1021/acs.biochem.6b00986
Single-Stranded Nucleic Acids Bind to the Tetramer Interface of SAMHD1 and Prevent Formation of the Catalytic Homotetramer
Abstract
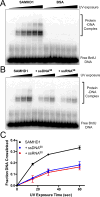
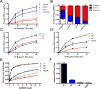
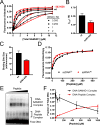
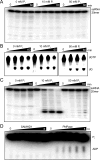
Sterile alpha motif and HD domain protein 1 (SAMHD1) is a unique enzyme that plays important roles in nucleic acid metabolism, viral restriction, and the pathogenesis of autoimmune diseases and cancer. Although much attention has been focused on its dNTP triphosphohydrolase activity in viral restriction and disease, SAMHD1 also binds to single-stranded RNA and DNA. Here we utilize a UV cross-linking method using 5-bromodeoxyuridine-substituted oligonucleotides coupled with high-resolution mass spectrometry to identify the binding site for single-stranded nucleic acids (ssNAs) on SAMHD1. Mapping cross-linked amino acids on the surface of existing crystal structures demonstrated that the ssNA binding site lies largely along the dimer-dimer interface, sterically blocking the formation of the homotetramer required for dNTPase activity. Surprisingly, the disordered C-terminus of SAMHD1 (residues 583-626) was also implicated in ssNA binding. An interaction between this region and ssNA was confirmed in binding studies using the purified SAMHD1 583-626 peptide. Despite a recent report that SAMHD1 possesses polyribonucleotide phosphorylase activity, we did not detect any such activity in the presence of inorganic phosphate, indicating that nucleic acid binding is unrelated to this proposed activity. These data suggest an antagonistic regulatory mechanism in which the mutually exclusive oligomeric state requirements for ssNA binding and dNTP hydrolase activity modulate these two functions of SAMHD1 within the cell.
Figures


References
-
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BCJ, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. - PMC - PubMed
-
- Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. - PubMed
-
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HCT, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LPS, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

