Stage-Wise Identification and Analysis of miRNA from Root-Knot Nematode Meloidogyne incognita
- PMID: 27775666
- PMCID: PMC5085782
- DOI: 10.3390/ijms17101758
Stage-Wise Identification and Analysis of miRNA from Root-Knot Nematode Meloidogyne incognita
Abstract
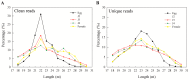
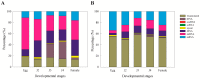
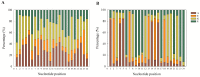
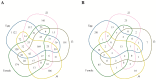
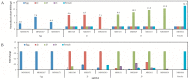
In this study, we investigated global changes in miRNAs of Meloidogyne incognita throughout its life cycle. Small RNA sequencing resulted in approximately 62, 38, 38, 35, and 39 Mb reads in the egg, J2, J3, J4, and female stages, respectively. Overall, we identified 2724 known and 383 novel miRNAs (read count > 10) from all stages, of which 169 known and 13 novel miRNA were common to all the five stages. Among the stage-specific miRNAs, miR-286 was highly expressed in eggs, miR-2401 in J2, miR-8 and miR-187 in J3, miR-6736 in J4, and miR-17 in the female stages. These miRNAs are reported to be involved in embryo and neural development, muscular function, and control of apoptosis. Cluster analysis indicated the presence of 91 miRNA clusters, of which 36 clusters were novel and identified in this study. Comparison of miRNA families with other nematodes showed 17 families to be commonly absent in animal parasitic nematodes and M. incognita. Validation of 43 predicted common and stage-specific miRNA by quantitative PCR (qPCR) indicated their expression in the nematode. Stage-wise exploration of M. incognita miRNAs has not been carried out before and this work presents information on common and stage-specific miRNAs of the root-knot nematode.
Keywords: Meloidogyne incognita; microRNA expression; quantitative polymerase chain reaction; stage-specific.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mitkowski N., Abawi G. Root-knot nematodes. Plant Health Instr. 2003 doi: 10.1094/PHI-I-2003-0917-01. - DOI
-
- Moens M., Perry R.N., Starr J.L. Meloidogyne Species—A Diverse Group of Novel and Important Plant Parasites. CABI; Wallingford, Oxfordshire, UK: 2009.
-
- Chitwood D.J. Nematicides. In: Plimmer J.R., editor. Encyclopedia Agrochemical. John Wiley & Sons; New York, NY, USA: 2003. pp. 1104–1115.
-
- Noling J. Movement and Toxicity of Nematicides in the Plant Root Zone. Department of Entomology and Nematology, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida; Gainesville, FL, USA: 2011. ENY-041 (formerly RF-NG002)
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

