RNA-binding protein CPEB1 remodels host and viral RNA landscapes
- PMID: 27775709
- PMCID: PMC5140759
- DOI: 10.1038/nsmb.3310
RNA-binding protein CPEB1 remodels host and viral RNA landscapes
Abstract
Host and virus interactions occurring at the post-transcriptional level are critical for infection but remain poorly understood. Here, we performed comprehensive transcriptome-wide analyses revealing that human cytomegalovirus (HCMV) infection results in widespread alternative splicing (AS), shortening of 3' untranslated regions (3' UTRs) and lengthening of poly(A)-tails in host gene transcripts. We found that the host RNA-binding protein CPEB1 was highly induced after infection, and ectopic expression of CPEB1 in noninfected cells recapitulated infection-related post-transcriptional changes. CPEB1 was also required for poly(A)-tail lengthening of viral RNAs important for productive infection. Strikingly, depletion of CPEB1 reversed infection-related cytopathology and post-transcriptional changes, and decreased productive HCMV titers. Host RNA processing was also altered in herpes simplex virus-2 (HSV-2)-infected cells, thereby indicating that this phenomenon might be a common occurrence during herpesvirus infections. We anticipate that our work may serve as a starting point for therapeutic targeting of host RNA-binding proteins in herpesvirus infections.
Figures


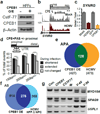


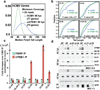
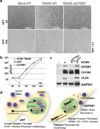
References
-
- Staras SA, et al. Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin Infect Dis. 2006;43:1143–1151. - PubMed
-
- Mocarski ES, et al. Betaherpes viral genes and their functions. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. 2007:204–230. - PubMed
-
- Fortunato EA, McElroy AK, Sanchez I, Spector DH. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 2000;8:111–119. - PubMed
-
- Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Howley DMKaPM., editor. Fields’ virology. 5th. Vol. 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2701–2772.
Methods References
-
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

