Mitochondrial genome sequences from wild and cultivated barley (Hordeum vulgare)
- PMID: 27776481
- PMCID: PMC5078923
- DOI: 10.1186/s12864-016-3159-3
Mitochondrial genome sequences from wild and cultivated barley (Hordeum vulgare)
Abstract
Background: Sequencing analysis of mitochondrial genomes is important for understanding the evolution and genome structures of various plant species. Barley is a self-pollinated diploid plant with seven chromosomes comprising a large haploid genome of 5.1 Gbp. Wild barley (Hordeum vulgare ssp. spontaneum) and cultivated barley (H. vulgare ssp. vulgare) have cross compatibility and closely related genomes, although a significant number of nucleotide polymorphisms have been reported between their genomes.
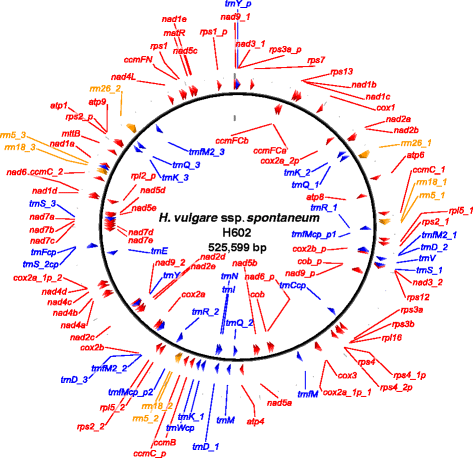
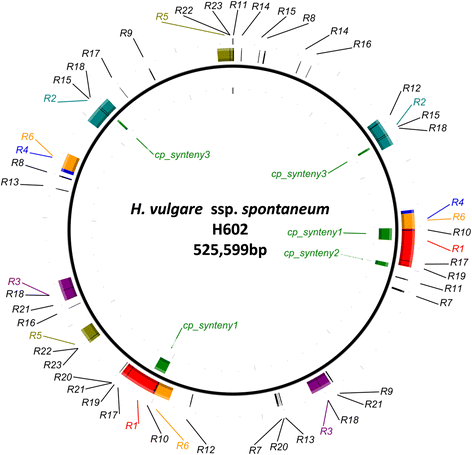
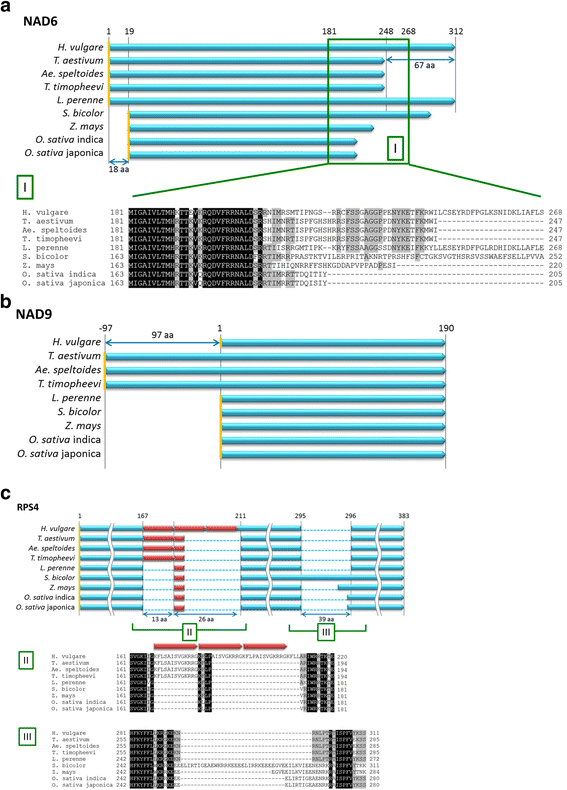
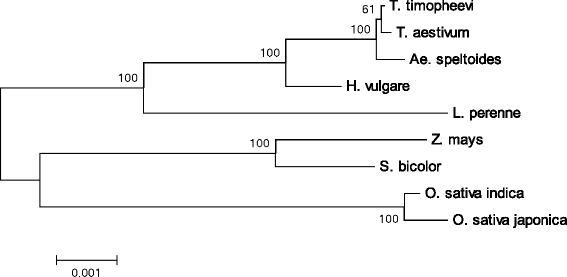
Results: We determined the complete nucleotide sequences of the mitochondrial genomes of wild and cultivated barley. Two independent circular maps of the 525,599 bp barley mitochondrial genome were constructed by de novo assembly of high-throughput sequencing reads of barley lines H602 and Haruna Nijo, with only three SNPs detected between haplotypes. These mitochondrial genomes contained 33 protein-coding genes, three ribosomal RNAs, 16 transfer RNAs, 188 new ORFs, six major repeat sequences and several types of transposable elements. Of the barley mitochondrial genome-encoded proteins, NAD6, NAD9 and RPS4 had unique structures among grass species.
Conclusions: The mitochondrial genome of barley was similar to those of other grass species in terms of gene content, but the configuration of the genes was highly differentiated from that of other grass species. Mitochondrial genome sequencing is essential for annotating the barley nuclear genome; our mitochondrial sequencing identified a significant number of fragmented mitochondrial sequences in the reported nuclear genome sequences. Little polymorphism was detected in the barley mitochondrial genome sequences, which should be explored further to elucidate the evolution of barley.
Keywords: Comparative genomics; De novo assembly; Hordeum vulgare; Mitochondrial genome.
Figures
Similar articles
-
History and future perspectives of barley genomics.DNA Res. 2020 Aug 1;27(4):dsaa023. doi: 10.1093/dnares/dsaa023. DNA Res. 2020. PMID: 32979265 Free PMC article. Review.
-
Chromosome-scale assembly of barley cv. 'Haruna Nijo' as a resource for barley genetics.DNA Res. 2022 Jan 28;29(1):dsac001. doi: 10.1093/dnares/dsac001. DNA Res. 2022. PMID: 35022669 Free PMC article.
-
Reference-based chromosome-scale assembly of Japanese barley (Hordeum vulgare ssp. vulgare) cultivar Hayakiso 2.DNA Res. 2025 Jul 4;32(4):dsaf016. doi: 10.1093/dnares/dsaf016. DNA Res. 2025. PMID: 40577690 Free PMC article.
-
Population genetics and phylogenetic analysis of the vrs1 nucleotide sequence in wild and cultivated barley.Genome. 2014 Apr;57(4):239-44. doi: 10.1139/gen-2014-0039. Epub 2014 Jun 27. Genome. 2014. PMID: 25033083
-
Prospects of pan-genomics in barley.Theor Appl Genet. 2019 Mar;132(3):785-796. doi: 10.1007/s00122-018-3234-z. Epub 2018 Nov 16. Theor Appl Genet. 2019. PMID: 30446793 Review.
Cited by
-
Deciphering the enigma of RNA editing in the ATP1_alpha subunit of ATP synthase in Triticum aestivum.Saudi J Biol Sci. 2023 Jul;30(7):103703. doi: 10.1016/j.sjbs.2023.103703. Epub 2023 Jun 9. Saudi J Biol Sci. 2023. PMID: 37389198 Free PMC article.
-
The mitochondrial genome of the diploid oat Avena longiglumis.BMC Plant Biol. 2023 Apr 26;23(1):218. doi: 10.1186/s12870-023-04217-8. BMC Plant Biol. 2023. PMID: 37098475 Free PMC article.
-
History and future perspectives of barley genomics.DNA Res. 2020 Aug 1;27(4):dsaa023. doi: 10.1093/dnares/dsaa023. DNA Res. 2020. PMID: 32979265 Free PMC article. Review.
-
Advancing the Conservation and Utilization of Barley Genetic Resources: Insights into Germplasm Management and Breeding for Sustainable Agriculture.Plants (Basel). 2023 Sep 6;12(18):3186. doi: 10.3390/plants12183186. Plants (Basel). 2023. PMID: 37765350 Free PMC article. Review.
-
Complete mitochondrial genome sequences of Brassica rapa (Chinese cabbage and mizuna), and intraspecific differentiation of cytoplasm in B. rapa and Brassica juncea.Breed Sci. 2017 Sep;67(4):357-362. doi: 10.1270/jsbbs.17023. Epub 2017 Jul 28. Breed Sci. 2017. PMID: 29085245 Free PMC article.
References
-
- Zohary D, Hopf E, Weiss M. Domestication of plants in the old world. 4th ed. Oxford University Press; 2012.
-
- Sato K, Flavell A, Russell J, Börner A, Valkoun J. Biotechnological approaches to barley improvement. In: Kumlehn J, Stein N, editors. Biotechnology in agriculture and forestry. Heidelberg: Springer; 2014. pp. 21–36.
-
- International Barley Sequencing Consortium (IBSC) A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–6. - PubMed
-
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci U S A. 2007;104:1424–9. doi: 10.1073/pnas.0608580104. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

