Infiltration-RNAseq: transcriptome profiling of Agrobacterium-mediated infiltration of transcription factors to discover gene function and expression networks in plants
- PMID: 27777610
- PMCID: PMC5069895
- DOI: 10.1186/s13007-016-0141-7
Infiltration-RNAseq: transcriptome profiling of Agrobacterium-mediated infiltration of transcription factors to discover gene function and expression networks in plants
Abstract
Background: Transcription factors (TFs) coordinate precise gene expression patterns that give rise to distinct phenotypic outputs. The identification of genes and transcriptional networks regulated by a TF often requires stable transformation and expression changes in plant cells. However, the production of stable transformants can be slow and laborious with no guarantee of success. Furthermore, transgenic plants overexpressing a TF of interest can present pleiotropic phenotypes and/or result in a high number of indirect gene expression changes. Therefore, fast, efficient, high-throughput methods for assaying TF function are needed.
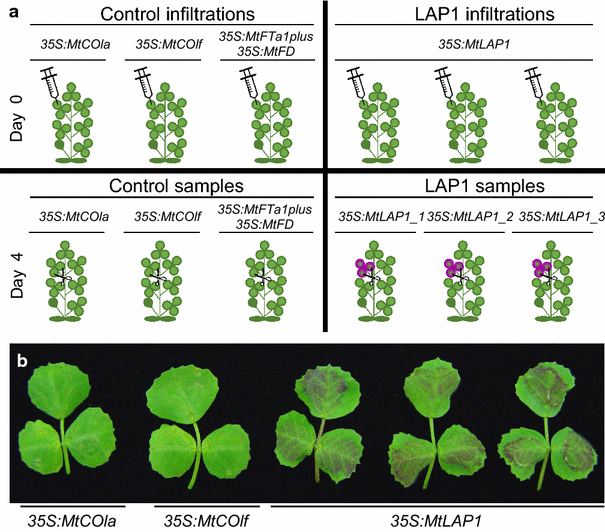
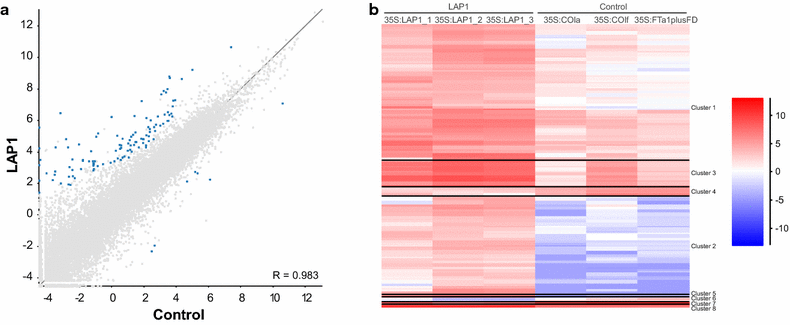
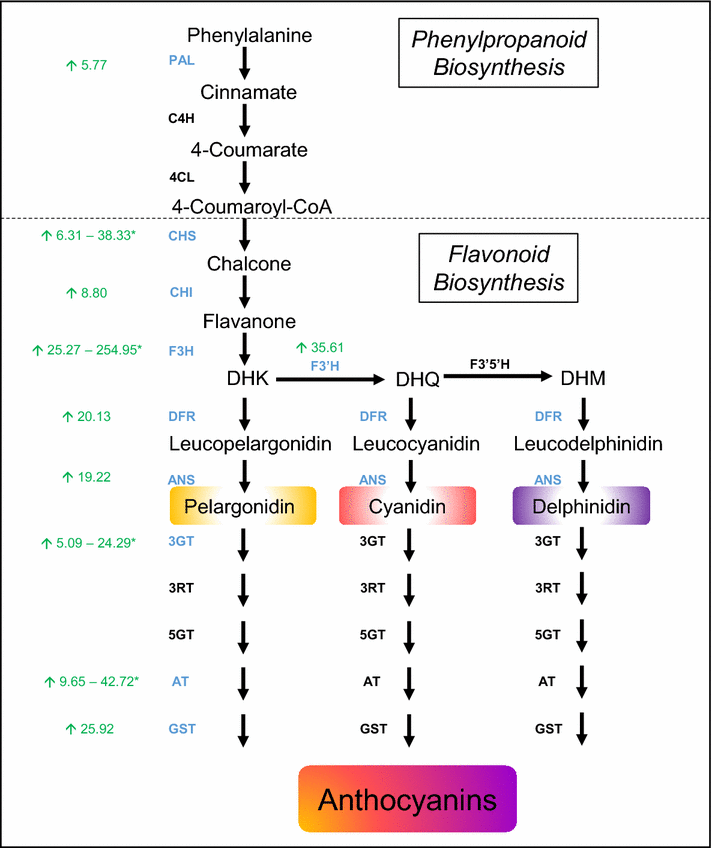
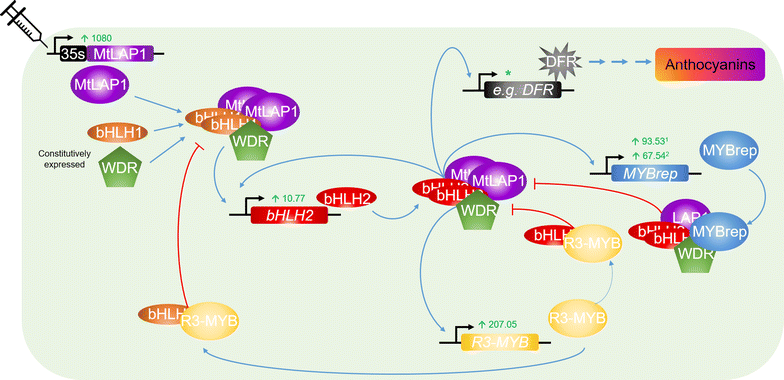
Results: Agroinfiltration is a simple plant biology method that allows transient gene expression. It is a rapid and powerful tool for the functional characterisation of TF genes in planta. High throughput RNA sequencing is now a widely used method for analysing gene expression profiles (transcriptomes). By coupling TF agroinfiltration with RNA sequencing (named here as Infiltration-RNAseq), gene expression networks and gene function can be identified within a few weeks rather than many months. As a proof of concept, we agroinfiltrated Medicago truncatula leaves with M. truncatula LEGUME ANTHOCYANIN PRODUCITION 1 (MtLAP1), a MYB transcription factor involved in the regulation of the anthocyanin pathway, and assessed the resulting transcriptome. Leaves infiltrated with MtLAP1 turned red indicating the production of anthocyanin pigment. Consistent with this, genes encoding enzymes in the anthocyanin biosynthetic pathway, and known transcriptional activators and repressors of the anthocyanin biosynthetic pathway, were upregulated. A novel observation was the induction of a R3-MYB transcriptional repressor that likely provides transcriptional feedback inhibition to prevent the deleterious effects of excess anthocyanins on photosynthesis.
Conclusions: Infiltration-RNAseq is a fast and convenient method for profiling TF-mediated gene expression changes. We utilised this method to identify TF-mediated transcriptional changes and TF target genes in M. truncatula and Nicotiana benthamiana. This included the identification of target genes of a TF not normally expressed in leaves, and targets of TFs from other plant species. Infiltration-RNAseq can be easily adapted to other plant species where agroinfiltration protocols have been optimised. The ability to identify downstream genes, including positive and negative transcriptional regulators, will result in a greater understanding of TF function.
Keywords: Agrobacterium tumefaciens; Anthocyanin; Infiltration; Medicago truncatula; MtLAP1; Nicotiana benthamiana; RNAseq; Transcription factor; Transcriptome.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

