Acquired resistance to innate immune clearance promotes Klebsiella pneumoniae ST258 pulmonary infection
- PMID: 27777978
- PMCID: PMC5070956
- DOI: 10.1172/jci.insight.89704
Acquired resistance to innate immune clearance promotes Klebsiella pneumoniae ST258 pulmonary infection
Abstract
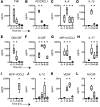
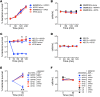
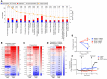
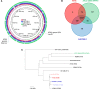
Adaptive changes in the genome of a locally predominant clinical isolate of the multidrug-resistant Klebsiella pneumoniae ST258 (KP35) were identified and help to explain the selection of this strain as a successful pulmonary pathogen. The acquisition of 4 new ortholog groups, including an arginine transporter, enabled KP35 to outcompete related ST258 strains lacking these genes. KP35 infection elicited a monocytic response, dominated by Ly6Chi monocytic myeloid-derived suppressor cells that lacked phagocytic capabilities, expressed IL-10, arginase, and antiinflammatory surface markers. In comparison with other K. pneumoniae strains, KP35 induced global changes in the phagocytic response identified with proteomics, including evasion of Ca2+ and calpain activation necessary for phagocytic killing, confirmed in functional studies with neutrophils. This comprehensive analysis of an ST258 K. pneumoniae isolate reveals ongoing genetic adaptation to host microenvironments and innate immune clearance mechanisms that complements its repertoire of antimicrobial resistance genes and facilitates persistence in the lung.
Figures



References
-
- Tumbarello M, et al. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother. 2015;70(7):2133–2143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

