Molecular Epidemiology of Colonizing and Infecting Isolates of Klebsiella pneumoniae
- PMID: 27777984
- PMCID: PMC5071533
- DOI: 10.1128/mSphere.00261-16
Molecular Epidemiology of Colonizing and Infecting Isolates of Klebsiella pneumoniae
Abstract
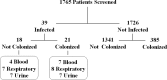
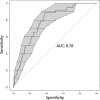
Klebsiella pneumoniae is among the most common causes of hospital-acquired infections and has emerged as an urgent threat to public health due to carbapenem antimicrobial resistance. K. pneumoniae commonly colonizes hospitalized patients and causes extraintestinal infections such as urinary tract infection, bloodstream infection (septicemia), and pneumonia. If colonization is an intermediate step before infection, then detection and characterization of colonizing isolates could enable strategies to prevent or empirically treat K. pneumoniae infections in hospitalized patients. However, the strength of the association between colonization and infection is unclear. To test the hypothesis that hospitalized patients become infected with their colonizing strain, 1,765 patients were screened for rectal colonization with K. pneumoniae, and extraintestinal isolates from these same patients were collected over a 3-month period in a cohort study design. The overall colonization prevalence was 23.0%. After adjustment for other patient factors, colonization was significantly associated with subsequent infection: 21 of 406 (5.2%) colonized patients later had extraintestinal infection, compared to 18 of 1,359 (1.3%) noncolonized patients (adjusted odds ratio [OR], 4.01; 95% confidence interval, 2.08 to 7.73; P < 0.001). Despite a high diversity of colonizing isolates, 7/7 respiratory, 4/4 urinary, and 2/5 bloodstream isolates from colonized patients matched the patient corresponding rectal swab isolates, based on wzi capsular typing, multilocus sequence typing (MLST), and whole-genome sequence analysis. These results suggest that K. pneumoniae colonization is directly associated with progression to extraintestinal infection. IMPORTANCE K. pneumoniae commonly infects hospitalized patients, and these infections are increasingly resistant to carbapenems, the antibiotics of last resort for life-threatening bacterial infections. To prevent and treat these infections, we must better understand how K. pneumoniae causes disease and discover new ways to predict and detect infections. This study demonstrates that colonization with K. pneumoniae in the intestinal tract is strongly linked to subsequent infection. This finding helps to identify a potential time frame and possible approach for intervention: the colonizing strain from a patient could be isolated as part of a risk assessment, and antibiotic susceptibility testing could guide empirical therapy if the patient becomes acutely ill.
Keywords: Klebsiella; MLST; cgMLST; colonization; infection; pneumonia; whole-genome sequencing; wzi.
Figures


Comment in
-
Genome watch: Klebsiella pneumoniae: when a colonizer turns bad.Nat Rev Microbiol. 2017 Jul;15(7):384. doi: 10.1038/nrmicro.2017.64. Epub 2017 Jun 5. Nat Rev Microbiol. 2017. PMID: 28579608 No abstract available.
References
-
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. - DOI - PMC - PubMed
-
- CDC 2014. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
