Racial differences in microRNA and gene expression in hypertensive women
- PMID: 27779208
- PMCID: PMC5078799
- DOI: 10.1038/srep35815
Racial differences in microRNA and gene expression in hypertensive women
Abstract
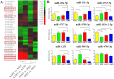
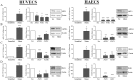
Systemic arterial hypertension is an important cause of cardiovascular disease morbidity and mortality. African Americans are disproportionately affected by hypertension, in fact the incidence, prevalence, and severity of hypertension is highest among African American (AA) women. Previous data suggests that differential gene expression influences individual susceptibility to selected diseases and we hypothesized that this phenomena may affect health disparities in hypertension. Transcriptional profiling of peripheral blood mononuclear cells from AA or white, normotensive or hypertensive females identified thousands of mRNAs differentially-expressed by race and/or hypertension. Predominant gene expression differences were observed in AA hypertensive females compared to AA normotensives or white hypertensives. Since microRNAs play important roles in regulating gene expression, we profiled global microRNA expression and observed differentially-expressed microRNAs by race and/or hypertension. We identified novel mRNA-microRNA pairs potentially involved in hypertension-related pathways and differently-expressed, including MCL1/miR-20a-5p, APOL3/miR-4763-5p, PLD1/miR-4717-3p, and PLD1/miR-4709-3p. We validated gene expression levels via RT-qPCR and microRNA target validation was performed in primary endothelial cells. Altogether, we identified significant gene expression differences between AA and white female hypertensives and pinpointed novel mRNA-microRNA pairs differentially-expressed by hypertension and race. These differences may contribute to the known disparities in hypertension and may be potential targets for intervention.
Figures




References
-
- National Heart, L., and Blood Institute Morbidity and Mortality: 2012 Chart Book on Cardiovascular, Lung, and Blood Diseases. (2012).
-
- In Health, United States, 2014: With Special Feature on Adults Aged 55-64 Health, United States (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

