ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions
- PMID: 27779234
- PMCID: PMC5078793
- DOI: 10.1038/srep35956
ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions
Abstract
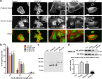
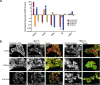
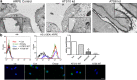
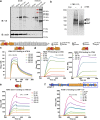
ADAMTS10 and ADAMTS6 are homologous metalloproteinases with ill-defined roles. ADAMTS10 mutations cause Weill-Marchesani syndrome (WMS), implicating it in fibrillin microfibril biology since some fibrillin-1 mutations also cause WMS. However little is known about ADAMTS6 function. ADAMTS10 is resistant to furin cleavage, however we show that ADAMTS6 is effectively processed and active. Using siRNA, over-expression and mutagenesis, it was found ADAMTS6 inhibits and ADAMTS10 is required for focal adhesions, epithelial cell-cell junction formation, and microfibril deposition. Either knockdown of ADAMTS6, or disruption of its furin processing or catalytic sites restores focal adhesions, implicating its enzyme activity acts on targets in the focal adhesion complex. In ADAMTS10-depleted cultures, expression of syndecan-4 rescues focal adhesions and cell-cell junctions. Recombinant C-termini of ADAMTS10 and ADAMTS6, both of which induce focal adhesions, bind heparin and syndecan-4. However, cells overexpressing full-length ADAMTS6 lack heparan sulphate and focal adhesions, whilst depletion of ADAMTS6 induces a prominent glycocalyx. Thus ADAMTS10 and ADAMTS6 oppositely affect heparan sulphate-rich interfaces including focal adhesions. We previously showed that microfibril deposition requires fibronectin-induced focal adhesions, and cell-cell junctions in epithelial cultures. Here we reveal that ADAMTS6 causes a reduction in heparan sulphate-rich interfaces, and its expression is regulated by ADAMTS10.
Figures



References
-
- Le Goff C. & Cormier-Daire V. From tall to short: the role of TGFbeta signaling in growth and its disorders. Am. J. Med. Genet. C Semin. Med. Genet. 160C, 145–153 (2012). - PubMed
-
- Somerville R. P., Jungers K. A. & Apte S. S. Discovery and characterization of a novel, widely expressed metalloprotease, ADAMTS10, and its proteolytic activation. J. Biol. Chem. 279, 51208–51217 (2004). - PubMed
-
- Baldwin A. K., Simpson A., Steer R., Cain S. A. & Kielty C. M. Elastic fibres in health and disease. Expert Rev. Mol. Med. 15, e8 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

