The global spectrum of protein-coding pharmacogenomic diversity
- PMID: 27779249
- PMCID: PMC5817389
- DOI: 10.1038/tpj.2016.77
The global spectrum of protein-coding pharmacogenomic diversity
Abstract
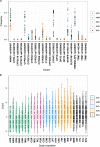
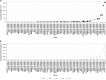
Differences in response to medications have a strong genetic component. By leveraging publically available data, the spectrum of such genomic variation can be investigated extensively. Pharmacogenomic variation was extracted from the 1000 Genomes Project Phase 3 data (2504 individuals, 26 global populations). A total of 12 084 genetic variants were found in 120 pharmacogenes, with the majority (90.0%) classified as rare variants (global minor allele frequency <0.5%), with 52.9% being singletons. Common variation clustered individuals into continental super-populations and 23 pharmacogenes contained highly differentiated variants (FST>0.5) for one or more super-population comparison. A median of three clinical variants (PharmGKB level 1A/B) was found per individual, and 55.4% of individuals carried loss-of-function variants, varying by super-population (East Asian 60.9%>African 60.1%>South Asian 60.3%>European 49.3%>Admixed 39.2%). Genome sequencing can therefore identify clinical pharmacogenomic variation, and future studies need to consider rare variation to understand the spectrum of genetic diversity contributing to drug response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Yip V, Hawcutt DB, Pirmohamed M. Pharmacogenetic markers of drug efficacy and toxicity. Clin Pharmacol Ther 2015; 98: 61–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

