Double-target Antisense U1snRNAs Correct Mis-splicing Due to c.639+861C>T and c.639+919G>A GLA Deep Intronic Mutations
- PMID: 27779620
- PMCID: PMC5095687
- DOI: 10.1038/mtna.2016.88
Double-target Antisense U1snRNAs Correct Mis-splicing Due to c.639+861C>T and c.639+919G>A GLA Deep Intronic Mutations
Abstract
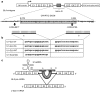
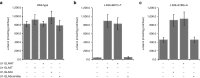
Fabry disease is a rare X-linked lysosomal storage disorder caused by deficiency of the α-galactosidase A (α-Gal A) enzyme, which is encoded by the GLA gene. GLA transcription in humans produces a major mRNA encoding α-Gal A and a minor mRNA of unknown function, which retains a 57-nucleotide-long cryptic exon between exons 4 and 5, bearing a premature termination codon. NM_000169.2:c.639+861C>T and NM_000169.2:c.639+919G>A GLA deep intronic mutations have been described to cause Fabry disease by inducing overexpression of the alternatively spliced mRNA, along with a dramatic decrease in the major one. Here, we built a wild-type GLA minigene and two minigenes that carry mutations c.639+861C>T and c.639+919G>A. Once transfected into cells, the minigenes recapitulate the molecular patterns observed in patients, at the mRNA, protein, and enzymatic level. We constructed a set of specific double-target U1asRNAs to correct c.639+861C>T and c.639+919G>A GLA mutations. Efficacy of U1asRNAs in inducing the skipping of the cryptic exon was evaluated upon their transient co-transfection with the minigenes in COS-1 cells, by real-time polymerase chain reaction (PCR), western blot analysis, and α-Gal A enzyme assay. We identified a set of U1asRNAs that efficiently restored α-Gal A enzyme activity and the correct splicing pathways in reporter minigenes. We also identified a unique U1asRNA correcting both mutations as efficently as the mutation-specific U1asRNAs. Our study proves that an exon skipping-based approach recovering α-Gal A activity in the c.639+861C>T and c.639+919G>A GLA mutations is active.
Figures


Similar articles
-
Identification and functional characterization of the first deep intronic GLA mutation (IVS4+1326C>T) causing renal variant of Fabry disease.Orphanet J Rare Dis. 2022 Jun 20;17(1):237. doi: 10.1186/s13023-022-02377-8. Orphanet J Rare Dis. 2022. PMID: 35725559 Free PMC article.
-
Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype.Am J Hum Genet. 2002 Apr;70(4):994-1002. doi: 10.1086/339431. Epub 2002 Feb 4. Am J Hum Genet. 2002. PMID: 11828341 Free PMC article.
-
Modulation the alternative splicing of GLA (IVS4+919G>A) in Fabry disease.PLoS One. 2017 Apr 21;12(4):e0175929. doi: 10.1371/journal.pone.0175929. eCollection 2017. PLoS One. 2017. PMID: 28430823 Free PMC article.
-
Identification of a Missense Mutation in the α-galactosidase A Gene in a Chinese Family with Fabry Disease.Curr Genomics. 2018 Jan;19(1):70-75. doi: 10.2174/1389202918666170915155033. Curr Genomics. 2018. PMID: 29491734 Free PMC article. Review.
-
The association of nonsense codons with exon skipping.Mutat Res. 1998 Sep;411(2):87-117. doi: 10.1016/s1383-5742(98)00010-6. Mutat Res. 1998. PMID: 9806422 Review.
Cited by
-
Transposon clusters as substrates for aberrant splice-site activation.RNA Biol. 2021 Mar;18(3):354-367. doi: 10.1080/15476286.2020.1805909. Epub 2020 Sep 23. RNA Biol. 2021. PMID: 32965162 Free PMC article.
-
Highlights on Genomics Applications for Lysosomal Storage Diseases.Cells. 2020 Aug 14;9(8):1902. doi: 10.3390/cells9081902. Cells. 2020. PMID: 32824006 Free PMC article. Review.
-
Long-read sequencing enables comprehensive molecular genetic diagnosis of Fabry disease.Hum Genomics. 2024 Nov 28;18(1):133. doi: 10.1186/s40246-024-00697-3. Hum Genomics. 2024. PMID: 39609713 Free PMC article.
-
Invention of an oral medication for cardiac Fabry disease caused by RNA mis-splicing.Sci Adv. 2025 Apr 11;11(15):eadt9695. doi: 10.1126/sciadv.adt9695. Epub 2025 Apr 9. Sci Adv. 2025. PMID: 40203112 Free PMC article.
-
Progressive myoclonus epilepsy in Gaucher Disease due to a new Gly-Gly mutation causing loss of an Exonic Splicing Enhancer.J Neurol. 2019 Jan;266(1):92-101. doi: 10.1007/s00415-018-9084-4. Epub 2018 Oct 31. J Neurol. 2019. PMID: 30382391 Free PMC article.
References
-
- Zarate, YA and Hopkin, RJ (2008). Fabry's disease. Lancet 372: 1427–1435. - PubMed
-
- Desnick, R, Ioannou, Y, and Eng, C (2001). α-Galactosidase a deficiency: Fabry disease. In: Scriver CR, Beaudet AL, Sly WS and Valle D (eds.). The Metabolic and Molecular Bases of Inherited Disease. 8th edn. McGraw-Hill: New York 2001: 3733–3774.
-
- Filoni, C, Caciotti, A, Carraresi, L, Donati, MA, Mignani, R, Parini, R et al. (2008). Unbalanced GLA mRNAs ratio quantified by real-time PCR in Fabry patients' fibroblasts results in Fabry disease. Eur J Hum Genet 16: 1311–1317. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

