Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology
- PMID: 27779741
- PMCID: PMC5290243
- DOI: 10.1111/imm.12684
Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology
Abstract
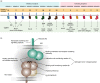
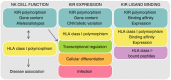
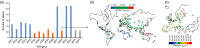
Killer-cell immunoglobulin-like receptors (KIRs) are components of two fundamental biological systems essential for human health and survival. First, they contribute to host immune responses, both innate and adaptive, through their expression by natural killer cells and T cells. Second, KIR play a key role in regulating placentation, and hence reproductive success. Analogous to the diversity of their human leucocyte antigen class I ligands, KIR are extremely polymorphic. In this review, we describe recent developments, fuelled by methodological advances, that are helping to decipher the KIR system in terms of haplotypes, polymorphisms, expression patterns and their ligand interactions. These developments are delivering deeper insight into the relevance of KIR in immune system function, evolution and disease.
Keywords: expression; haplotypes; killer-cell immunoglobulin-like receptors; ligands; natural killer cell; polymorphism.
© 2016 The Authors. Immunology Published by John Wiley & Sons Ltd.
Figures






References
-
- Battistini L, Borsellino G, Sawicki G, Poccia F, Salvetti M, Ristori G et al Phenotypic and cytokine analysis of human peripheral blood γδ T cells expressing NK cell receptors. J Immunol 1997; 159:3723–30. - PubMed
-
- Young NT, Uhrberg M, Phillips JH, Lanier LL, Parham P. Differential expression of leukocyte receptor complex‐encoded Ig‐like receptors correlates with the transition from effector to memory CTL. J Immunol 2001; 166:3933–41. - PubMed
-
- Patterson S, Chaidos A, Neville DC, Poggi A, Butters TD, Roberts IA et al Human invariant NKT cells display alloreactivity instructed by invariant TCR‐CD1d interaction and killer Ig receptors. J Immunol 2008; 181:3268–76. - PubMed
-
- van Bergen J, Kooy‐Winkelaar EM, van Dongen H, van Gaalen FA, Thompson A, Huizinga TW et al Functional killer Ig‐like receptors on human memory CD4+ T cells specific for cytomegalovirus. J Immunol 2009; 182:4175–82. - PubMed
-
- Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D et al Coordinated expression of Ig‐like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol 2004; 173:7223–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

