Developmental and Thyroid Hormone Regulation of the DNA Methyltransferase 3a Gene in Xenopus Tadpoles
- PMID: 27779916
- PMCID: PMC5133355
- DOI: 10.1210/en.2016-1465
Developmental and Thyroid Hormone Regulation of the DNA Methyltransferase 3a Gene in Xenopus Tadpoles
Abstract
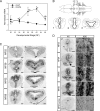
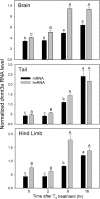
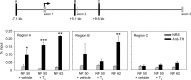
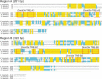
Thyroid hormone is essential for normal development in vertebrates. In amphibians, T3 controls metamorphosis by inducing tissue-specific gene regulation programs. A hallmark of T3 action is the modification of chromatin structure, which underlies changes in gene transcription. We found that mRNA for the de novo DNA methyltransferase (DNMT) dnmt3a, but not dnmt1, increased in the brain of Xenopus tadpoles during metamorphosis in parallel with plasma [T3]. Addition of T3 to the rearing water caused a time-dependent increase in dnmt3a mRNA in tadpole brain, tail, and hind limb. By analyzing data from a genome-wide analysis of T3 receptor (TR) binding in tadpole tail, we identified several putative T3 response elements (TREs) within the dnmt3a locus. Using in vitro DNA binding, transient transfection-reporter, and chromatin immunoprecipitation assays for TRs, we identified two functional TREs at -7.1 kb and +5.1 kb relative to the dnmt3a transcription start site. Sequence alignment showed that these TREs are conserved between two related frog species, X. laevis and X. tropicalis, but not with amniotes. Our previous findings showed that this gene is directly regulated by liganded TRs in mouse brain, and whereas the two mouse TREs are conserved among Eutherian mammals, they are not conserved in Xenopus species. Thus, although T3 regulation of dnmt3a may be an ancient pathway in vertebrates, the genomic sites responsible for hormone regulation may have diverged or arisen by convergent evolution. We hypothesize that direct T3 regulation of dnmt3a may be an important mechanism for modulating global changes in DNA methylation.
Figures

References
-
- Bernal J. Action of thyroid hormone in brain. J Endocrinol Invest. 2002;25(3):268–288. - PubMed
-
- Anderson GW, Schoonover CM, Jones SA. Control of thyroid hormone action in the developing rat brain. Thyroid. 2003;13(11):1039–1056. - PubMed
-
- Gudernatsch JF. Feeding experiments on tadpoles. I. The influence of specific organs given as food on growth and differentiation. A contribution to the knowledge of organs with internal secretion. Wilhelm Roux Arch Entwickl Mech Org. 1912;35(3):457–483.
-
- Denver RJ. Neuroendocrinology of amphibian metamorphosis. In: Shi YB, ed. Current Topics in Developmental Biology: Animal Metamorphosis. Vol 103 Chap 7 San Diego, CA: Elsevier; 2013:195–227. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

